RegTransBase--a database of regulatory sequences and interactions based on literature: a resource for investigating transcriptional regulation in prokaryotes
- PMID: 23547897
- PMCID: PMC3639892
- DOI: 10.1186/1471-2164-14-213
RegTransBase--a database of regulatory sequences and interactions based on literature: a resource for investigating transcriptional regulation in prokaryotes
Abstract
Background: Due to the constantly growing number of sequenced microbial genomes, comparative genomics has been playing a major role in the investigation of regulatory interactions in bacteria. Regulon inference mostly remains a field of semi-manual examination since absence of a knowledgebase and informatics platform for automated and systematic investigation restricts opportunities for computational prediction. Additionally, confirming computationally inferred regulons by experimental data is critically important.
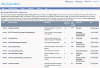
Description: RegTransBase is an open-access platform with a user-friendly web interface publicly available at http://regtransbase.lbl.gov. It consists of two databases - a manually collected hierarchical regulatory interactions database based on more than 7000 scientific papers which can serve as a knowledgebase for verification of predictions, and a large set of curated by experts transcription factor binding sites used in regulon inference by a variety of tools. RegTransBase captures the knowledge from published scientific literature using controlled vocabularies and contains various types of experimental data, such as: the activation or repression of transcription by an identified direct regulator; determination of the transcriptional regulatory function of a protein (or RNA) directly binding to DNA or RNA; mapping of binding sites for a regulatory protein; characterization of regulatory mutations. Analysis of the data collected from literature resulted in the creation of Putative Regulons from Experimental Data that are also available in RegTransBase.
Conclusions: RegTransBase is a powerful user-friendly platform for the investigation of regulation in prokaryotes. It uses a collection of validated regulatory sequences that can be easily extracted and used to infer regulatory interactions by comparative genomics techniques thus assisting researchers in the interpretation of transcriptional regulation data.
Figures


References
-
- Rodionov DA, Novichkov PS, Stavrovskaya ED, Rodionova IA, Li X, Kazanov MD, Ravcheev DA, Gerasimova AV, Kazakov AE, Kovaleva GY. Comparative genomic reconstruction of transcriptional networks controlling central metabolism in the Shewanella genus. BMC Genomics. 2011;12(Suppl 1):S3. doi: 10.1186/1471-2164-12-S1-S3. - DOI - PMC - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources

