CSAR benchmark exercise 2011-2012: evaluation of results from docking and relative ranking of blinded congeneric series
- PMID: 23548044
- PMCID: PMC3753884
- DOI: 10.1021/ci400025f
CSAR benchmark exercise 2011-2012: evaluation of results from docking and relative ranking of blinded congeneric series
Abstract
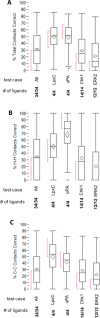
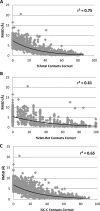
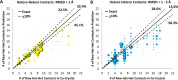
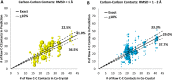
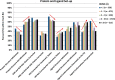
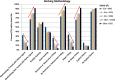
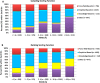
The Community Structure-Activity Resource (CSAR) recently held its first blinded exercise based on data provided by Abbott, Vertex, and colleagues at the University of Michigan, Ann Arbor. A total of 20 research groups submitted results for the benchmark exercise where the goal was to compare different improvements for pose prediction, enrichment, and relative ranking of congeneric series of compounds. The exercise was built around blinded high-quality experimental data from four protein targets: LpxC, Urokinase, Chk1, and Erk2. Pose prediction proved to be the most straightforward task, and most methods were able to successfully reproduce binding poses when the crystal structure employed was co-crystallized with a ligand from the same chemical series. Multiple evaluation metrics were examined, and we found that RMSD and native contact metrics together provide a robust evaluation of the predicted poses. It was notable that most scoring functions underpredicted contacts between the hetero atoms (i.e., N, O, S, etc.) of the protein and ligand. Relative ranking was found to be the most difficult area for the methods, but many of the scoring functions were able to properly identify Urokinase actives from the inactives in the series. Lastly, we found that minimizing the protein and correcting histidine tautomeric states positively trended with low RMSD for pose prediction but minimizing the ligand negatively trended. Pregenerated ligand conformations performed better than those that were generated on the fly. Optimizing docking parameters and pretraining with the native ligand had a positive effect on the docking performance as did using restraints, substructure fitting, and shape fitting. Lastly, for both sampling and ranking scoring functions, the use of the empirical scoring function appeared to trend positively with the RMSD. Here, by combining the results of many methods, we hope to provide a statistically relevant evaluation and elucidate specific shortcomings of docking methodology for the community.
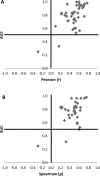
Figures




References
-
- Jorgensen W. L. The many roles of computation in drug discovery. Science 2004, 303, 1813–1818. - PubMed
-
- Leach A. R.; Shoichet B. K.; Peishoff C. E. Prediction of protein–ligand interactions. Docking and scoring: Successes and gaps. J. Med. Chem. 2006, 49, 5851–5855. - PubMed
-
- Lyne P. D. Structure-based virtual screening: An overview. Drug. Discovery Today 2002, 7, 1047–1055. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Miscellaneous

