Donor DNA Utilization During Gene Targeting with Zinc-Finger Nucleases
- PMID: 23550125
- PMCID: PMC3618352
- DOI: 10.1534/g3.112.005439
Donor DNA Utilization During Gene Targeting with Zinc-Finger Nucleases
Abstract
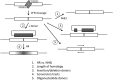
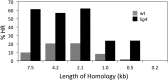
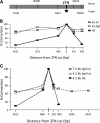
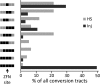
Gene targeting is the term commonly applied to experimental gene replacement by homologous recombination (HR). This process is substantially stimulated by a double-strand break (DSB) in the genomic target. Zinc-finger nucleases (ZFNs) are targetable cleavage reagents that provide an effective means of introducing such a break in conjunction with delivery of a homologous donor DNA. In this study we explored several parameters of donor DNA structure during ZFN-mediated gene targeting in Drosophila melanogaster embryos, as follows. 1) We confirmed that HR outcomes are enhanced relative to the alternative nonhomologous end joining (NHEJ) repair pathway in flies lacking DNA ligase IV. 2) The minimum amount of homology needed to support efficient HR in fly embryos is between 200 and 500 bp. 3) Conversion tracts are very broad in this system: donor sequences more than 3 kb from the ZFN-induced break are found in the HR products at approximately 50% of the frequency of a marker at the break. 4) Deletions carried by the donor DNA are readily incorporated at the target. 5) While linear double-stranded DNAs are not effective as donors, single-stranded oligonucleotides are. These observations should enable better experimental design for gene targeting in Drosophila and help guide similar efforts in other systems.
Keywords: Gene targeting; conversion tracts; homologous recombination; nonhomologous end joining; zinc-finger nucleases.
Copyright © 2013 Beumer et al.
Figures


References
-
- Bibikova M., Beumer K., Trautman J. K., Carroll D., 2003. Enhancing gene targeting with designed zinc finger nucleases. Science 300: 764. - PubMed
Publication types
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials
