Activation-Induced Cytidine Deaminase Does Not Impact Murine Meiotic Recombination
- PMID: 23550130
- PMCID: PMC3618351
- DOI: 10.1534/g3.113.005553
Activation-Induced Cytidine Deaminase Does Not Impact Murine Meiotic Recombination
Erratum in
-
Corrigendum.G3 (Bethesda). 2015 Oct 5;5(10):2217. doi: 10.1534/g3.115.020917. G3 (Bethesda). 2015. PMID: 26438608 Free PMC article. No abstract available.
Abstract
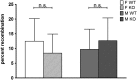
Activation-induced cytidine deaminase (AID) was first described as the triggering enzyme of the B-cell-specific reactions that edit the immunoglobulin genes, namely somatic hypermutation, gene conversion, and class switch recombination. Over the years, AID was also detected in cells other than lymphocytes, and it has been assigned additional roles in the innate defense against transforming retroviruses, in retrotransposition restriction and in DNA demethylation. Notably, AID expression was found in germline tissues, and in heterologous systems it can induce the double-strand breaks required for the initiation of meiotic recombination and proper gamete formation. However, because AID-deficient mice are fully fertile, the molecule is not essential for meiosis. Thus, the remaining question that we addressed here is whether AID influences the frequency of meiotic recombination in mice. We measured the recombination events in the meiosis of male and female mice F1 hybrids of C57BL/6J and BALB/c, in Aicda+/+ and Aicda-/- background by using a panel of single-nucleotide polymorphisms that distinguishes C57BL/6J from BALB/c genome across the 19 autosomes. In agreement with the literature, we found that the frequency of recombination in the female germline was greater than in male germline, both in the Aicda+/+ and Aicda-/- backgrounds. No statistical difference was found in the average recombination events between Aicda+/+ and Aidca-/- animals, either in females or males. In addition, the recombination frequencies between single-nucleotide polymorphisms flanking the immunoglobulin heavy and immunoglobulin kappa loci was also not different. We conclude that AID has a minor impact, if any, on the overall frequency of meiotic recombination.
Keywords: AID; cytidine deaminase; double-strand breaks; germline; meiotic recombination.
Copyright © 2013 Cortesao et al.
Figures


Comment on
-
Activation-induced cytidine deaminase deaminates 5-methylcytosine in DNA and is expressed in pluripotent tissues: implications for epigenetic reprogramming.J Biol Chem. 2004 Dec 10;279(50):52353-60. doi: 10.1074/jbc.M407695200. Epub 2004 Sep 24. J Biol Chem. 2004. PMID: 15448152
-
Alternative induction of meiotic recombination from single-base lesions of DNA deaminases.Genetics. 2009 May;182(1):41-54. doi: 10.1534/genetics.109.101683. Epub 2009 Feb 23. Genetics. 2009. PMID: 19237686 Free PMC article.
References
-
- Arakawa H., Hauschild J., Buerstedde J.-M., 2002. Requirement of the activation-induced deaminase (AID) gene for immunoglobulin gene conversion. Science 295: 1301–1306. - PubMed
-
- Barreto V. M., Magor B. G., 2011. Activation-induced cytidine deaminase structure and functions: a species comparative view. Dev. Comp. Immunol. 35: 991–1007. - PubMed
-
- Bastos H., Lassalle B., Chicheportiche A., Riou L., Testart J., et al. , 2005. Flow cytometric characterization of viable meiotic and postmeiotic cells by Hoechst 33342 in mouse spermatogenesis. Cytometry A 65: 40–49. - PubMed
-
- Baudat F., Manova K., Yuen J. P., Jasin M., Keeney S., 2000. Chromosome synapsis defects and sexually dimorphic meiotic progression in mice lacking Spo11. Mol. Cell 6: 989–998. - PubMed
-
- Begum N. A., Honjo T., 2012. Evolutionary comparison of the mechanism of DNA cleavage with respect to immune diversity and genomic instability. Biochemistry 51: 5243–5256. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
