Trans-Lesion DNA Polymerases May Be Involved in Yeast Meiosis
- PMID: 23550131
- PMCID: PMC3618350
- DOI: 10.1534/g3.113.005603
Trans-Lesion DNA Polymerases May Be Involved in Yeast Meiosis
Abstract
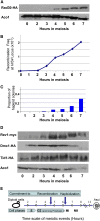
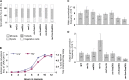
Trans-lesion DNA polymerases (TLSPs) enable bypass of DNA lesions during replication and are also induced under stress conditions. Being only weakly dependent on their template during replication, TLSPs introduce mutations into DNA. The low processivity of these enzymes ensures that they fall off their template after a few bases are synthesized and are then replaced by the more accurate replicative polymerase. We find that the three TLSPs of budding yeast Saccharomyces cerevisiae Rev1, PolZeta (Rev3 and Rev7), and Rad30 are induced during meiosis at a time when DNA double-strand breaks (DSBs) are formed and homologous chromosomes recombine. Strains deleted for one or any combination of the three TLSPs undergo normal meiosis. However, in the triple-deletion mutant, there is a reduction in both allelic and ectopic recombination. We suggest that trans-lesion polymerases are involved in the processing of meiotic double-strand breaks that lead to mutations. In support of this notion, we report significant yeast two-hybrid (Y2H) associations in meiosis-arrested cells between the TLSPs and DSB proteins Rev1-Spo11, Rev1-Mei4, and Rev7-Rec114, as well as between Rev1 and Rad30 We suggest that the involvement of TLSPs in processing of meiotic DSBs could be responsible for the considerably higher frequency of mutations reported during meiosis compared with that found in mitotically dividing cells, and therefore may contribute to faster evolutionary divergence than previously assumed.
Keywords: DNA repair; DSB processing; meiosis; recombination; trans-lesion synthesis polymerases.
Copyright © 2013 by the Genetics Society of America.
Figures


References
-
- Arora C., Kee K., Maleki S., Keeney S., 2004. Antiviral protein Ski8 is a direct partner of Spo11 in meiotic DNA break formation, independent of its cytoplasmic role in RNA metabolism. Mol. Cell 13: 549–559 - PubMed
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Miscellaneous
