Comparative Genomics of Serial Isolates of Cryptococcus neoformans Reveals Gene Associated With Carbon Utilization and Virulence
- PMID: 23550133
- PMCID: PMC3618354
- DOI: 10.1534/g3.113.005660
Comparative Genomics of Serial Isolates of Cryptococcus neoformans Reveals Gene Associated With Carbon Utilization and Virulence
Abstract
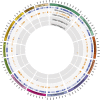
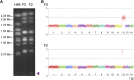
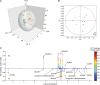
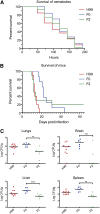
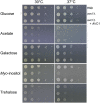
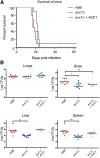
The opportunistic fungal pathogen Cryptococcus neoformans is a leading cause of mortality among the human immunodeficiency virus/acquired immunodeficiency syndrome population and is known for frequently causing life-threatening relapses. To investigate the potential contribution of in-host microevolution to persistence and relapse, we have analyzed two serial isolates obtained from a patient with acquired immunodeficiency syndrome who suffered an initial and relapse episode of cryptococcal meningoencephalitis. Despite being identical by multilocus sequence typing, the isolates differ phenotypically, exhibiting changes in key virulence factors, nutrient acquisition, metabolic profiles, and the ability to disseminate in an animal model. Whole-genome sequencing uncovered a clonal relationship, with only a few unique differences. Of these, two key changes are expected to explain the phenotypic differences observed in the relapse isolate: loss of a predicted AT-rich interaction domain protein and changes in copy number of the left and right arms of chromosome 12. Gene deletion of the predicted transcriptional regulator produced changes in melanin, capsule, carbon source use, and dissemination in the host, consistent with the phenotype of the relapse isolate. In addition, the deletion mutant displayed altered virulence in the murine model. The observed differences suggest the relapse isolate evolved subsequent to penetration of the central nervous system and may have gained dominance following the administration of antifungal therapy. These data reveal the first molecular insights into how the Cryptococcus neoformans genome changes during infection of humans and the manner in which microevolution progresses in this deadly fungal pathogen.
Keywords: cryptococcosis; fungal pathogenesis; microevolution; relapse.
Copyright © 2013 Ormerod et al.
Figures



References
-
- Altschul S. F., Gish W., Miller W., Myers E. W., Lipman D. J., 1990. Basic local alignment search tool. J. Mol. Biol. 215: 403–410 - PubMed
-
- Bubb W. A., Wright L. C., Cagney M., Santangelo R. T., Sorrell T. C., et al. , 1999. Heteronuclear NMR studies of metabolites produced by Cryptococcus neoformans in culture media: Identification of possible virulence factors. Magn. Reson. Med. 42: 442–453 - PubMed
Publication types
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
