Microbe-independent entry of oomycete RxLR effectors and fungal RxLR-like effectors into plant and animal cells is specific and reproducible
- PMID: 23550528
- PMCID: PMC3994703
- DOI: 10.1094/MPMI-02-13-0051-IA
Microbe-independent entry of oomycete RxLR effectors and fungal RxLR-like effectors into plant and animal cells is specific and reproducible
Abstract
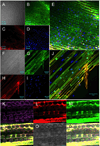
A wide diversity of pathogens and mutualists of plant and animal hosts, including oomycetes and fungi, produce effector proteins that enter the cytoplasm of host cells. A major question has been whether or not entry by these effectors can occur independently of the microbe or requires machinery provided by the microbe. Numerous publications have documented that oomycete RxLR effectors and fungal RxLR-like effectors can enter plant and animal cells independent of the microbe. A recent reexamination of whether the RxLR domain of oomycete RxLR effectors is sufficient for microbe-independent entry into host cells concluded that the RxLR domains of Phytophthora infestans Avr3a and of P. sojae Avr1b alone are NOT sufficient to enable microbe-independent entry of proteins into host and nonhost plant and animal cells. Here, we present new, more detailed data that unambiguously demonstrate that the RxLR domain of Avr1b does show efficient and specific entry into soybean root cells and also into wheat leaf cells, at levels well above background nonspecific entry. We also summarize host cell entry experiments with a wide diversity of oomycete and fungal effectors with RxLR or RxLR-like motifs that have been independently carried out by the seven different labs that coauthored this letter. Finally we discuss possible technical reasons why specific cell entry may have been not detected by Wawra et al. (2013).
Figures

References
-
- Anderson RG, Casady MS, Fee RA, Vaughan MM, Deb D, Fedkenheuer K, Huffaker A, Schmelz EA, Tyler BM, McDowell JM. Homologous RXLR effectors from Hyaloperonospora arabidopsidis and Phytophthora sojae suppress immunity in distantly related plants. Plant J. 2012;72:882–893. - PubMed
-
- Boothroyd JC, Dubremetz JF. Kiss and spit: the dual roles of Toxoplasma rhoptries. Nat Rev Microbiol. 2008;6:79–88. - PubMed
-
- Chang M, Chou JC, Lee HJ. Cellular internalization of fluorescent proteins via arginine-rich intracellular delivery peptide in plant cells. Plant & cell physiology. 2005;46:482–488. - PubMed
-
- Ellis J, Catanzariti AM, Dodds P. The problem of how fungal and oomycete avirulence proteins enter plant cells. Trends Plant Sci. 2006;11:61–63. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

