MicroRNA regulation of T-cell differentiation and function
- PMID: 23550639
- PMCID: PMC3621017
- DOI: 10.1111/imr.12061
MicroRNA regulation of T-cell differentiation and function
Abstract
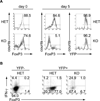
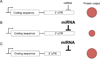
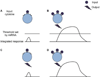
MicroRNAs (miRNAs) are emerging as key controllers of T-cell differentiation and function. Their expression is dynamically regulated by extracellular signals such as costimulation and cytokine signals. miRNAs set thresholds for gene expression and optimize protein concentrations of genetic networks. Absence of individual miRNAs can lead to severe immune dysfunction. In this study, we review emerging principles and provide examples of important functions exerted by miRNAs. Although our understanding of miRNA function in T-cell differentiation is still rudimentary, the available evidence leaves no doubt that these small post-transcriptional regulators are indispensable for proper functioning of the immune system.
© 2013 John Wiley & Sons A/S. Published by Blackwell Publishing Ltd.
Conflict of interest statement
The authors have no conflicts of interest to declare.
Figures


References
-
- Sharp PA. The centrality of RNA. Cell. 2009;136:577–580. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

