Control of alternative splicing in immune responses: many regulators, many predictions, much still to learn
- PMID: 23550649
- PMCID: PMC3621013
- DOI: 10.1111/imr.12047
Control of alternative splicing in immune responses: many regulators, many predictions, much still to learn
Abstract
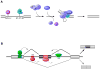
Most mammalian pre-mRNAs are alternatively spliced in a manner that alters the resulting open reading frame. Consequently, alternative pre-mRNA splicing provides an important RNA-based layer of protein regulation and cellular function. The ubiquitous nature of alternative splicing coupled with the advent of technologies that allow global interrogation of the transcriptome have led to an increasing awareness of the possibility that widespread changes in splicing patterns contribute to lymphocyte function during an immune response. Indeed, a few notable examples of alternative splicing have clearly been demonstrated to regulate T-cell responses to antigen. Moreover, several proteins key to the regulation of splicing in T cells have recently been identified. However, much remains to be done to truly identify the spectrum of genes that are regulated at the level of splicing in immune cells and to determine how many of these are controlled by currently known factors and pathways versus unknown mechanisms. Here, we describe the proteins, pathways, and mechanisms that have been shown to regulate alternative splicing in human T cells and discuss what is and is not known about the genes regulated by such factors. Finally, we highlight unifying themes with regards to the mechanisms and consequences of alternative splicing in the adaptive immune system and give our view of important directions for future studies.
© 2013 John Wiley & Sons A/S. Published by Blackwell Publishing Ltd.
Conflict of interest statement
The authors have no conflicts of interest to declare.
Figures


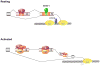

References
-
- Moore MJ, Proudfoot NJ. Pre-mRNA processing reaches back to transcription and ahead to translation. Cell. 2009;136:688–700. - PubMed
-
- Pan Q, Shai O, Lee LJ, Frey BJ, Blencowe BJ. Deep surveying of alternative splicing complexity in the human transcriptome by high-throughput sequencing. Nat Genet. 2008;40:1413–1415. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

