Different distributions of Epstein-Barr virus early and late gene transcripts within viral replication compartments
- PMID: 23552415
- PMCID: PMC3676136
- DOI: 10.1128/JVI.00219-13
Different distributions of Epstein-Barr virus early and late gene transcripts within viral replication compartments
Abstract
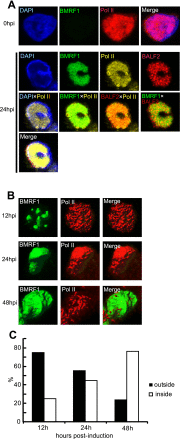
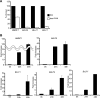
Productive replication of the Epstein-Barr virus (EBV) occurs in discrete sites in nuclei, called replication compartments, where viral genome DNA synthesis and transcription take place. The replication compartments include subnuclear domains, designated BMRF1 cores, which are highly enriched in the BMRF1 protein. During viral lytic replication, newly synthesized viral DNA genomes are organized around and then stored inside BMRF1 cores. Here, we examined spatial distribution of viral early and late gene mRNAs within replication compartments using confocal laser scanning microscopy and three-dimensional surface reconstruction imaging. EBV early mRNAs were mainly located outside the BMRF1 cores, while viral late mRNAs were identified inside, corresponding well with the fact that late gene transcription is dependent on viral DNA replication. From these results, we speculate that sites for viral early and late gene transcription are separated with reference to BMRF1 cores.
Figures


Similar articles
-
Spatiotemporally different DNA repair systems participate in Epstein-Barr virus genome maturation.J Virol. 2011 Jul;85(13):6127-35. doi: 10.1128/JVI.00258-11. Epub 2011 Apr 13. J Virol. 2011. PMID: 21490093 Free PMC article.
-
Epstein-Barr virus genome packaging factors accumulate in BMRF1-cores within viral replication compartments.PLoS One. 2019 Sep 13;14(9):e0222519. doi: 10.1371/journal.pone.0222519. eCollection 2019. PLoS One. 2019. PMID: 31518362 Free PMC article.
-
The SWI/SNF Chromatin Regulator BRG1 Modulates the Transcriptional Regulatory Activity of the Epstein-Barr Virus DNA Polymerase Processivity Factor BMRF1.J Virol. 2017 Apr 13;91(9):e02114-16. doi: 10.1128/JVI.02114-16. Print 2017 May 1. J Virol. 2017. PMID: 28228591 Free PMC article.
-
Latent and lytic Epstein-Barr virus replication strategies.Rev Med Virol. 2005 Jan-Feb;15(1):3-15. doi: 10.1002/rmv.441. Rev Med Virol. 2005. PMID: 15386591 Review.
-
Replication Compartments-The Great Survival Strategy for Epstein-Barr Virus Lytic Replication.Microorganisms. 2022 Apr 25;10(5):896. doi: 10.3390/microorganisms10050896. Microorganisms. 2022. PMID: 35630341 Free PMC article. Review.
Cited by
-
Differential carbonic anhydrase activities control EBV-induced B-cell transformation and lytic cycle reactivation.PLoS Pathog. 2024 Mar 26;20(3):e1011998. doi: 10.1371/journal.ppat.1011998. eCollection 2024 Mar. PLoS Pathog. 2024. PMID: 38530845 Free PMC article.
-
Genomic characterization of HIV-associated plasmablastic lymphoma identifies pervasive mutations in the JAK-STAT pathway.Blood Cancer Discov. 2020 Jul;1(1):112-125. doi: 10.1158/2643-3230.BCD-20-0051. Blood Cancer Discov. 2020. PMID: 33225311 Free PMC article.
-
Continuous DNA replication is required for late gene transcription and maintenance of replication compartments in gammaherpesviruses.PLoS Pathog. 2018 May 29;14(5):e1007070. doi: 10.1371/journal.ppat.1007070. eCollection 2018 May. PLoS Pathog. 2018. PMID: 29813138 Free PMC article.
-
Direct Evidence of Abortive Lytic Infection-Mediated Establishment of Epstein-Barr Virus Latency During B-Cell Infection.Front Microbiol. 2021 Jan 21;11:575255. doi: 10.3389/fmicb.2020.575255. eCollection 2020. Front Microbiol. 2021. PMID: 33613459 Free PMC article.
-
Epigenetic lifestyle of Epstein-Barr virus.Semin Immunopathol. 2020 Apr;42(2):131-142. doi: 10.1007/s00281-020-00792-2. Epub 2020 Mar 30. Semin Immunopathol. 2020. PMID: 32232535 Free PMC article. Review.
References
-
- Baer R, Bankier AT, Biggin MD, Deininger PL, Farrell PJ, Gibson TJ, Hatfull G, Hudson GS, Satchwell SC, Seguin C, et al. 1984. DNA sequence and expression of the B95-8 Epstein-Barr virus genome. Nature 310:207–211 - PubMed
-
- Babcock GJ, Decker LL, Volk M, Thorley-Lawson DA. 1998. EBV persistence in memory B cells in vivo. Immunity 9:395–404 - PubMed
-
- Henle W. 1972. Role of Epstein-Barr virus in infectious mononucleosis and malignant lymphomas in man. Fed. Proc. 31:1674. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources

