Human La protein interaction with GCAC near the initiator AUG enhances hepatitis C Virus RNA replication by promoting linkage between 5' and 3' untranslated regions
- PMID: 23552417
- PMCID: PMC3676093
- DOI: 10.1128/JVI.00525-13
Human La protein interaction with GCAC near the initiator AUG enhances hepatitis C Virus RNA replication by promoting linkage between 5' and 3' untranslated regions
Abstract
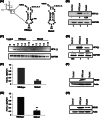
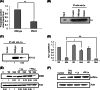
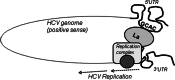
Human La protein has been implicated in facilitating the internal initiation of translation as well as replication of hepatitis C virus (HCV) RNA. Previously, we demonstrated that La interacts with the HCV internal ribosome entry site (IRES) around the GCAC motif near the initiator AUG within stem-loop IV by its RNA recognition motif (RRM) (residues 112 to 184) and influences HCV translation. In this study, we have deciphered the role of this interaction in HCV replication in a hepatocellular carcinoma cell culture system. We incorporated mutation of the GCAC motif in an HCV monocistronic subgenomic replicon and a pJFH1 construct which altered the binding of La and checked HCV RNA replication by reverse transcriptase PCR (RT-PCR). The mutation drastically affected HCV replication. Furthermore, to address whether the decrease in replication is a consequence of translation inhibition or not, we incorporated the same mutation into a bicistronic replicon and observed a substantial decrease in HCV RNA levels. Interestingly, La overexpression rescued this inhibition of replication. More importantly, we observed that the mutation reduced the association between La and NS5B. The effect of the GCAC mutation on the translation-to-replication switch, which is regulated by the interplay between NS3 and La, was further investigated. Additionally, our analyses of point mutations in the GCAC motif revealed distinct roles of each nucleotide in HCV replication and translation. Finally, we showed that a specific interaction of the GCAC motif with human La protein is crucial for linking 5' and 3' ends of the HCV genome. Taken together, our results demonstrate the mechanism of regulation of HCV replication by interaction of the cis-acting element GCAC within the HCV IRES with human La protein.
Figures








Similar articles
-
La protein binding at the GCAC site near the initiator AUG facilitates the ribosomal assembly on the hepatitis C virus RNA to influence internal ribosome entry site-mediated translation.J Biol Chem. 2004 Jul 16;279(29):29879-88. doi: 10.1074/jbc.M403417200. Epub 2004 May 10. J Biol Chem. 2004. PMID: 15138264
-
Structural determinant of human La protein critical for internal initiation of translation of hepatitis C virus RNA.J Virol. 2008 Dec;82(23):11927-38. doi: 10.1128/JVI.00924-08. Epub 2008 Oct 1. J Virol. 2008. PMID: 18829760 Free PMC article.
-
Human La antigen is required for the hepatitis C virus internal ribosome entry site-mediated translation.J Biol Chem. 2000 Sep 8;275(36):27531-40. doi: 10.1074/jbc.M001487200. J Biol Chem. 2000. PMID: 10856291
-
Hepatitis C Virus Translation Regulation.Int J Mol Sci. 2020 Mar 27;21(7):2328. doi: 10.3390/ijms21072328. Int J Mol Sci. 2020. PMID: 32230899 Free PMC article. Review.
-
cis-Acting RNA elements in the hepatitis C virus RNA genome.Virus Res. 2015 Aug 3;206:90-8. doi: 10.1016/j.virusres.2014.12.029. Epub 2015 Jan 7. Virus Res. 2015. PMID: 25576644 Free PMC article. Review.
Cited by
-
Nuclear proteins hijacked by mammalian cytoplasmic plus strand RNA viruses.Virology. 2015 May;479-480:457-74. doi: 10.1016/j.virol.2015.03.001. Epub 2015 Mar 26. Virology. 2015. PMID: 25818028 Free PMC article. Review.
-
HuR Displaces Polypyrimidine Tract Binding Protein To Facilitate La Binding to the 3' Untranslated Region and Enhances Hepatitis C Virus Replication.J Virol. 2015 Nov;89(22):11356-71. doi: 10.1128/JVI.01714-15. Epub 2015 Sep 2. J Virol. 2015. PMID: 26339049 Free PMC article.
-
Untranslated Regions of a Segmented Kindia Tick Virus Genome Are Highly Conserved and Contain Multiple Regulatory Elements for Viral Replication.Microorganisms. 2024 Jan 23;12(2):239. doi: 10.3390/microorganisms12020239. Microorganisms. 2024. PMID: 38399643 Free PMC article.
-
XRN1 stalling in the 5' UTR of Hepatitis C virus and Bovine Viral Diarrhea virus is associated with dysregulated host mRNA stability.PLoS Pathog. 2015 Mar 6;11(3):e1004708. doi: 10.1371/journal.ppat.1004708. eCollection 2015 Mar. PLoS Pathog. 2015. PMID: 25747802 Free PMC article.
-
Hydrogen peroxide induces La cytoplasmic shuttling and increases hepatitis C virus internal ribosome entry site-dependent translation.J Gen Virol. 2016 Sep;97(9):2301-2315. doi: 10.1099/jgv.0.000556. Epub 2016 Jul 18. J Gen Virol. 2016. PMID: 27436793 Free PMC article.
References
-
- Suzuki T, Aizaki H, Murakami K, Shoji I, Wakita T. 2007. Molecular biology of hepatitis C virus. J. Gastroenterol. 42:411–423 - PubMed
-
- Suzuki T, Ishii K, Aizaki H, Wakita T. 2007. Hepatitis C viral life cycle. Adv. Drug Deliv. Rev. 59:1200–1212 - PubMed
-
- Choo QL, Kuo G, Weiner AJ, Overby LR, Bradley DW, Houghton M. 1989. Isolation of a cDNA clone derived from a blood-borne non-A, non-B viral hepatitis genome. Science 244:359–362 - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

