Selection of a highly invasive neuroblastoma cell population through long-term human cytomegalovirus infection
- PMID: 23552602
- PMCID: PMC3412641
- DOI: 10.1038/oncsis.2012.10
Selection of a highly invasive neuroblastoma cell population through long-term human cytomegalovirus infection
Abstract
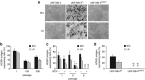
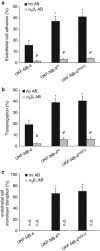
The human cytomegalovirus (HCMV) is suspected to increase tumour malignancy by infection of cancer and/or stroma cells (oncomodulation). So far, oncomodulatory mechanisms have been attributed to the presence of HCMV and direct action of its gene products on cancer cells. Here, we investigated whether the prolonged presence of HCMV can result in the irreversible selection of a cancer cell population with increased malignancy. The neuroblastoma cell line UKF-NB-4 was long-term (200 passages) infected with the HCMV strain Hi91 (UKF-NB-4(Hi)) before virus eradication using ganciclovir (UKF-NB-4(HiGCV)). Global gene expression profiling of UKF-NB-4, UKF-NB-4(Hi) and UKF-NB-4(HiGCV) cells and subsequent bioinformatic signal transduction pathway analysis revealed clear differences between UKF-NB-4 and UKF-NB-4(Hi), as well as between UKF-NB-4 and UKF-NB-4(HiGCV) cells, but only minor differences between UKF-NB-4(Hi) and UKF-NB-4(HiGCV) cells. Investigation of the expression of a subset of five genes in different chronically HCMV-infected cell lines before and after virus eradication suggested that long-term HCMV infection reproducibly causes specific changes. Array comparative genomic hybridisation showed virtually the same genomic differences for the comparisons UKF-NB-4(Hi)/UKF-NB-4 and UKF-NB-4(HiGCV)/UKF-NB-4. UKF-NB-4(Hi) cells are characterised by an increased invasive potential compared with UKF-NB-4 cells. This phenotype was completely retained in UKF-NB-4(HiGCV) cells. Moreover, there was a substantial overlap in the signal transduction pathways that differed significantly between UKF-NB-4(Hi)/UKF-NB-4(HiGCV) and UKF-NB-4 cells and those differentially regulated between tumour tissues from neuroblastoma patients with favourable or poor outcome. In conclusion, we present the first experimental evidence that long-term HCMV infection can result in the selection of tumour cell populations with enhanced malignancy.
Figures

References
-
- Söderberg-Nauclér C. HCMV microinfections in inflammatory diseases and cancer. J Clin Virol. 2008;41:218–223. - PubMed
-
- Michaelis M, Baumgarten P, Mittelbronn M, Hernáiz Driever P, Doerr HW, Cinatl J. Oncomodulation by human cytomegalovirus: Novel clinical findings open new roads. Med Microbiol Immunol. 2011;200:1–5. - PubMed
-
- Cinatl J, Cinatl J, Radsak K, Rabenau H, Weber B, Novak M, et al. Replication of human cytomegalovirus in a rhabdomyosarcoma cell line depends on the state of differentiation of the cells. Arch Virol. 1994;138:391–401. - PubMed
LinkOut - more resources
Full Text Sources

