The CEACAM1 tumor suppressor is an ATM and p53-regulated gene required for the induction of cellular senescence by DNA damage
- PMID: 23552604
- PMCID: PMC3412640
- DOI: 10.1038/oncsis.2012.7
The CEACAM1 tumor suppressor is an ATM and p53-regulated gene required for the induction of cellular senescence by DNA damage
Abstract
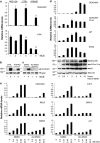
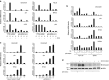
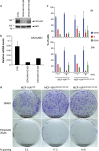
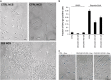
The p53 tumor-suppressor protein has a key role in the induction of cellular senescence, an important barrier to cancer development. However, very little is known about the physiological mediators of cellular senescence induced by p53. CEACAM1 is an immunoglobulin superfamily member whose expression is frequently lost in human tumors and exhibits tumor-suppressor features in several experimental systems, including Ceacam1 knockout mice. There is currently little understanding of the pathways and mechanisms by which CEACAM1 exerts its tumor-suppressor function. Here we report that CEACAM1 is strongly upregulated during the cellular response to DNA double-strand breaks (DSBs) starting from the lowest doses of DSB inducers used, and that upregulation is mediated by the ataxia telangiectasia mutated (ATM)/p53 pathway. Stable silencing of CEACAM1 showed that CEACAM1 is required for p53-mediated cellular senescence, but not initial cell growth arrest, in response to DNA damage. These findings identify CEACAM1 as a key component of the ATM/p53-mediated cellular response to DNA damage, and as a tumor suppressor mediating cellular senescence downstream of p53.
Figures

Similar articles
-
Chromium (VI) activates ataxia telangiectasia mutated (ATM) protein. Requirement of ATM for both apoptosis and recovery from terminal growth arrest.J Biol Chem. 2003 May 16;278(20):17885-94. doi: 10.1074/jbc.M210560200. Epub 2003 Mar 10. J Biol Chem. 2003. PMID: 12637545
-
Functional interaction of H2AX, NBS1, and p53 in ATM-dependent DNA damage responses and tumor suppression.Mol Cell Biol. 2005 Jan;25(2):661-70. doi: 10.1128/MCB.25.2.661-670.2005. Mol Cell Biol. 2005. PMID: 15632067 Free PMC article.
-
ATM: the protein encoded by the gene mutated in the radiosensitive syndrome ataxia-telangiectasia.Int J Radiat Biol. 1999 Oct;75(10):1201-14. doi: 10.1080/095530099139359. Int J Radiat Biol. 1999. PMID: 10549596 Review.
-
Ataxia telangiectasia mutated (ATM)-mediated DNA damage response in oxidative stress-induced vascular endothelial cell senescence.J Biol Chem. 2010 Sep 17;285(38):29662-70. doi: 10.1074/jbc.M110.125138. Epub 2010 Jul 16. J Biol Chem. 2010. PMID: 20639198 Free PMC article.
-
The influence of heterochromatin on DNA double strand break repair: Getting the strong, silent type to relax.DNA Repair (Amst). 2010 Dec 10;9(12):1273-82. doi: 10.1016/j.dnarep.2010.09.013. Epub 2010 Oct 30. DNA Repair (Amst). 2010. PMID: 21036673 Review.
Cited by
-
Condurango-glycoside-A fraction of Gonolobus condurango induces DNA damage associated senescence and apoptosis via ROS-dependent p53 signalling pathway in HeLa cells.Mol Cell Biochem. 2013 Oct;382(1-2):173-83. doi: 10.1007/s11010-013-1732-5. Epub 2013 Jun 27. Mol Cell Biochem. 2013. PMID: 23807740
-
A549 cells adapted to high nitric oxide show reduced surface CEACAM expression and altered adhesion and migration properties.Tumour Biol. 2015 Mar;36(3):1871-9. doi: 10.1007/s13277-014-2789-9. Epub 2014 Dec 11. Tumour Biol. 2015. PMID: 25500969
-
Glycosylation Alters Dimerization Properties of a Cell-surface Signaling Protein, Carcinoembryonic Antigen-related Cell Adhesion Molecule 1 (CEACAM1).J Biol Chem. 2016 Sep 16;291(38):20085-95. doi: 10.1074/jbc.M116.740050. Epub 2016 Jul 28. J Biol Chem. 2016. PMID: 27471271 Free PMC article.
-
Tumor intrinsic immunity related proteins may be novel tumor suppressors in some types of cancer.Sci Rep. 2019 Jul 29;9(1):10918. doi: 10.1038/s41598-019-47382-3. Sci Rep. 2019. PMID: 31358815 Free PMC article.
-
Dissecting phenotypic transitions in metastatic disease via photoconversion-based isolation.Elife. 2021 Feb 23;10:e63270. doi: 10.7554/eLife.63270. Elife. 2021. PMID: 33620315 Free PMC article.
References
-
- Nouvion AL, Beauchemin N. CEACAM1 as a central modulator of metabolism, tumor progression, angiogenesis and immunity] Med Sci. 2009;25:247–252. - PubMed
-
- Renwick A, Thompson D, Seal S, Kelly P, Chagtai T, Ahmed M, et al. ATM mutations that cause ataxia-telangiectasia are breast cancer susceptibility alleles. Nat Genet. 2006;38:873–875. - PubMed
-
- Gorgoulis VG, Halazonetis TD. Oncogene-induced senescence: the bright and dark side of the response. Curr Opin Cell Biol. 2010;22:816–827. - PubMed
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Research Materials
Miscellaneous

