The Drosophila F-box protein dSkp2 regulates cell proliferation by targeting Dacapo for degradation
- PMID: 23552694
- PMCID: PMC3667721
- DOI: 10.1091/mbc.E12-10-0772
The Drosophila F-box protein dSkp2 regulates cell proliferation by targeting Dacapo for degradation
Abstract
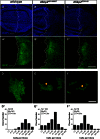
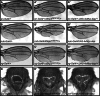
Cell cycle progression is controlled by a complex regulatory network consisting of interacting positive and negative factors. In humans, the positive regulator Skp2, an F-box protein, has been a subject of intense investigation in part because of its oncogenic activity. By contrast, the molecular and developmental functions of its Drosophila homologue, dSkp2, are poorly understood. Here we investigate the role of dSkp2 by focusing on its functional relationship with Dacapo (Dap), the Drosophila homologue of the cyclin-dependent kinase inhibitors p21(cip1)/p27(kip1)/p57(kip2). We show that dSkp2 interacts physically with Dap and has a role in targeting Dap for ubiquitination and proteasome-mediated degradation. We present evidence that dSkp2 regulates cell cycle progression by antagonizing Dap in vivo. dSkp2 knockdown reduces cell density in the wing by prolonging the cell doubling time. In addition, the wing phenotype caused by dSkp2 knockdown resembles that caused by dap overexpression and can be partially suppressed by reducing the gene dose of dap. Our study thus documents a conserved functional relationship between dSkp2 and Dap in their control of cell cycle progression, suggesting the possibility of using Drosophila as a model system to study Skp2-mediated tumorigenesis.
Figures





Similar articles
-
Dicer-1-dependent Dacapo suppression acts downstream of Insulin receptor in regulating cell division of Drosophila germline stem cells.Development. 2009 May;136(9):1497-507. doi: 10.1242/dev.025999. Epub 2009 Mar 31. Development. 2009. PMID: 19336466 Free PMC article.
-
The NUCKS1-SKP2-p21/p27 axis controls S phase entry.Nat Commun. 2021 Nov 29;12(1):6959. doi: 10.1038/s41467-021-27124-8. Nat Commun. 2021. PMID: 34845229 Free PMC article.
-
The Cyclin-dependent kinase inhibitor Dacapo promotes genomic stability during premeiotic S phase.Mol Biol Cell. 2009 Apr;20(7):1960-9. doi: 10.1091/mbc.e08-09-0916. Epub 2009 Feb 11. Mol Biol Cell. 2009. PMID: 19211840 Free PMC article.
-
The role of Skp2 and its substrate CDKN1B (p27) in colorectal cancer.J Gastrointestin Liver Dis. 2015 Jun;24(2):225-34. doi: 10.15403/jgld.2014.1121.242.skp2. J Gastrointestin Liver Dis. 2015. PMID: 26114183 Review.
-
p27(Kip1) signaling: Transcriptional and post-translational regulation.Int J Biochem Cell Biol. 2015 Nov;68:9-14. doi: 10.1016/j.biocel.2015.08.005. Epub 2015 Aug 14. Int J Biochem Cell Biol. 2015. PMID: 26279144 Review.
Cited by
-
Robustness of cell cycle control and flexible orders of signaling events.Sci Rep. 2015 Sep 30;5:14627. doi: 10.1038/srep14627. Sci Rep. 2015. PMID: 26419873 Free PMC article.
-
Expression of an S phase-stabilized version of the CDK inhibitor Dacapo can alter endoreplication.Development. 2015 Dec 15;142(24):4288-98. doi: 10.1242/dev.115006. Epub 2015 Oct 22. Development. 2015. PMID: 26493402 Free PMC article.
-
Various applications of TALEN- and CRISPR/Cas9-mediated homologous recombination to modify the Drosophila genome.Biol Open. 2014 Apr 15;3(4):271-80. doi: 10.1242/bio.20147682. Biol Open. 2014. PMID: 24659249 Free PMC article.
-
SCF(Slmb) E3 ligase-mediated degradation of Expanded is inhibited by the Hippo pathway in Drosophila.Cell Res. 2015 Jan;25(1):93-109. doi: 10.1038/cr.2014.166. Epub 2014 Dec 19. Cell Res. 2015. PMID: 25522691 Free PMC article.
References
-
- Amati B, Vlach J. Kip1 meets SKP2: new links in cell-cycle control. Nat Cell Biol. 1999;1:E91–E93. - PubMed
-
- Ben-Izhak O, Lahav-Baratz S, Meretyk S, Ben-Eliezer S, Sabo E, Dirnfeld M, Cohen S, Ciechanover A. Inverse relationship between Skp2 ubiquitin ligase and the cyclin dependent kinase inhibitor p27Kip1 in prostate cancer. J Urol. 2003;170:241–245. - PubMed
-
- Bhattacharya S, Garriga J, Calbo J, Yong T, Haines DS, Grana X. SKP2 associates with p130 and accelerates p130 ubiquitylation and degradation in human cells. Oncogene. 2003;22:2443–2451. - PubMed
-
- Bond M, Sala-Newby GB, Wu YJ, Newby AC. Biphasic effect of p21Cip1 on smooth muscle cell proliferation: role of PI 3-kinase and Skp2-mediated degradation. Cardiovasc Res. 2006;69:198–206. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials
Miscellaneous

