A decade and a half of protein intrinsic disorder: biology still waits for physics
- PMID: 23553817
- PMCID: PMC3690711
- DOI: 10.1002/pro.2261
A decade and a half of protein intrinsic disorder: biology still waits for physics
Abstract
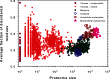
The abundant existence of proteins and regions that possess specific functions without being uniquely folded into unique 3D structures has become accepted by a significant number of protein scientists. Sequences of these intrinsically disordered proteins (IDPs) and IDP regions (IDPRs) are characterized by a number of specific features, such as low overall hydrophobicity and high net charge which makes these proteins predictable. IDPs/IDPRs possess large hydrodynamic volumes, low contents of ordered secondary structure, and are characterized by high structural heterogeneity. They are very flexible, but some may undergo disorder to order transitions in the presence of natural ligands. The degree of these structural rearrangements varies over a very wide range. IDPs/IDPRs are tightly controlled under the normal conditions and have numerous specific functions that complement functions of ordered proteins and domains. When lacking proper control, they have multiple roles in pathogenesis of various human diseases. Gaining structural and functional information about these proteins is a challenge, since they do not typically "freeze" while their "pictures are taken." However, despite or perhaps because of the experimental challenges, these fuzzy objects with fuzzy structures and fuzzy functions are among the most interesting targets for modern protein research. This review briefly summarizes some of the recent advances in this exciting field and considers some of the basic lessons learned from the analysis of physics, chemistry, and biology of IDPs.
© 2013 The Protein Society.
Figures


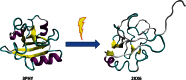

References
-
- Fischer E. Einfluss der configuration auf die wirkung der enzyme. Ber Dt Chem Ges. 1894;27:2985–2993.
-
- Bernstein FC, Koetzle TF, Williams GJ, Meyer EF, Jr, Brice MD, Rodgers JR, Kennard O, Shimanouchi T, Tasumi M. The Protein Data Bank: a computer-based archival file for macromolecular structures. J Mol Biol. 1977;112:535–542. - PubMed
-
- Kuhn TS. The structure of scientific revolutions. Chicago: University Of Chicago Press; 2012.
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources

