Funnel metadynamics as accurate binding free-energy method
- PMID: 23553839
- PMCID: PMC3631651
- DOI: 10.1073/pnas.1303186110
Funnel metadynamics as accurate binding free-energy method
Abstract
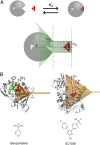
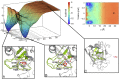
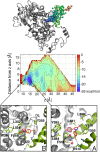
A detailed description of the events ruling ligand/protein interaction and an accurate estimation of the drug affinity to its target is of great help in speeding drug discovery strategies. We have developed a metadynamics-based approach, named funnel metadynamics, that allows the ligand to enhance the sampling of the target binding sites and its solvated states. This method leads to an efficient characterization of the binding free-energy surface and an accurate calculation of the absolute protein-ligand binding free energy. We illustrate our protocol in two systems, benzamidine/trypsin and SC-558/cyclooxygenase 2. In both cases, the X-ray conformation has been found as the lowest free-energy pose, and the computed protein-ligand binding free energy in good agreement with experiments. Furthermore, funnel metadynamics unveils important information about the binding process, such as the presence of alternative binding modes and the role of waters. The results achieved at an affordable computational cost make funnel metadynamics a valuable method for drug discovery and for dealing with a variety of problems in chemistry, physics, and material science.
Conflict of interest statement
The authors declare no conflict of interest.
Figures
References
-
- Jorgensen WL. The many roles of computation in drug discovery. Science. 2004;303(5665):1813–1818. - PubMed
-
- Gilson MK, Zhou HX. Calculation of protein–ligand binding affinities. Annu Rev Biophys Biomol Struct. 2007;36:21–42. - PubMed
-
- Goodsell DS, Olson AJ. Automated docking of substrates to proteins by simulated annealing. Proteins. 1990;8(3):195–202. - PubMed
-
- Meng EC, Shoichet BK, Kuntz ID. Automated docking with grid-based energy evaluation. J Comput Chem. 1992;13(4):505–524.
-
- Abagyan RA, Totrov MM, Kuznetsov DA. ICM—A new method for protein modeling and design: Applications to docking and structure prediction from the distorted native conformation. J Comput Chem. 1994;15(5):488–506.
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Research Materials

