Identification of Gram-positive cocci by use of matrix-assisted laser desorption ionization-time of flight mass spectrometry: comparison of different preparation methods and implementation of a practical algorithm for routine diagnostics
- PMID: 23554198
- PMCID: PMC3716085
- DOI: 10.1128/JCM.02654-12
Identification of Gram-positive cocci by use of matrix-assisted laser desorption ionization-time of flight mass spectrometry: comparison of different preparation methods and implementation of a practical algorithm for routine diagnostics
Abstract
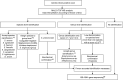
This study compared three sample preparation methods (direct transfer, the direct transfer-formic acid method with on-target formic acid treatment, and ethanol-formic acid extraction) for the identification of Gram-positive cocci with matrix-assisted laser desorption ionization-time of flight mass spectrometry (MALDI-TOF MS). A total of 156 Gram-positive cocci representing the clinically most important genera, Aerococcus, Enterococcus, Staphylococcus, and Streptococcus, as well as more rare genera, such as Gemella and Granulicatella, were analyzed using a Bruker MALDI Biotyper. The rate of correct genus-level identifications was approximately 99% for all three sample preparation methods. The species identification rate was significantly higher for the direct transfer-formic acid method and ethanol-formic acid extraction (both 77.6%) than for direct transfer (64.1%). Using direct transfer-formic acid compared to direct transfer, the total time to result was increased by 22.6%, 16.4%, and 8.5% analyzing 12, 48, and 96 samples per run, respectively. In a subsequent prospective study, 1,619 clinical isolates of Gram-positive cocci were analyzed under routine conditions by MALDI-TOF MS, using the direct transfer-formic acid preparation, and by conventional biochemical methods. For 95.6% of the isolates, a congruence between conventional and MALDI-TOF MS identification was observed. Two major limitations were found using MALDI-TOF MS: the differentiation of members of the Streptococcus mitis group and the identification of Streptococcus dysgalactiae. The Bruker MALDI Biotyper system using the direct transfer-formic acid sample preparation method was shown to be a highly reliable tool for the identification of Gram-positive cocci. We here suggest a practical algorithm for the clinical laboratory combining MALDI-TOF MS with phenotypic and molecular methods.
Figures
References
-
- Cherkaoui A, Hibbs J, Emonet S, Tangomo M, Girard M, François P, Schrenzel J. 2010. Comparison of two matrix-assisted laser desorption ionization-time of flight mass spectrometry methods with conventional phenotypic identification for routine identification of bacteria to the species level. J. Clin. Microbiol. 48:1169–1175 - PMC - PubMed
Publication types
MeSH terms
LinkOut - more resources
Full Text Sources
Other Literature Sources

