Antimicrobial resistance and virulence: a successful or deleterious association in the bacterial world?
- PMID: 23554414
- PMCID: PMC3623377
- DOI: 10.1128/CMR.00059-12
Antimicrobial resistance and virulence: a successful or deleterious association in the bacterial world?
Abstract
Hosts and bacteria have coevolved over millions of years, during which pathogenic bacteria have modified their virulence mechanisms to adapt to host defense systems. Although the spread of pathogens has been hindered by the discovery and widespread use of antimicrobial agents, antimicrobial resistance has increased globally. The emergence of resistant bacteria has accelerated in recent years, mainly as a result of increased selective pressure. However, although antimicrobial resistance and bacterial virulence have developed on different timescales, they share some common characteristics. This review considers how bacterial virulence and fitness are affected by antibiotic resistance and also how the relationship between virulence and resistance is affected by different genetic mechanisms (e.g., coselection and compensatory mutations) and by the most prevalent global responses. The interplay between these factors and the associated biological costs depend on four main factors: the bacterial species involved, virulence and resistance mechanisms, the ecological niche, and the host. The development of new strategies involving new antimicrobials or nonantimicrobial compounds and of novel diagnostic methods that focus on high-risk clones and rapid tests to detect virulence markers may help to resolve the increasing problem of the association between virulence and resistance, which is becoming more beneficial for pathogenic bacteria.
Figures
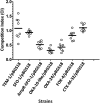
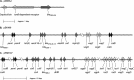
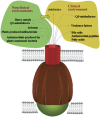
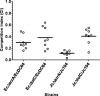




References
-
- Madigan MT, Martinko JM, Dunlap PV, Clark DP. 2009. Brock biology of microorganisms, 12th ed Pearson Education Inc., Upper Saddle River, NJ
-
- Feldman MW, Laland KN. 1996. Gene-culture coevolutionary theory. Trends Ecol. Evol. 11:453–457 - PubMed
-
- Burrus V, Waldor MK. 2004. Shaping bacterial genomes with integrative and conjugative elements. Res. Microbiol. 155:376–386 - PubMed
-
- Handel A, Regoes RR, Antia R. 2006. The role of compensatory mutations in the emergence of drug resistance. PLoS Comput. Biol. 2:e137 doi:10.1371/journal.pcbi.0020137 - DOI - PMC - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical

