Transcription factor-microRNA-target gene networks associated with ovarian cancer survival and recurrence
- PMID: 23554906
- PMCID: PMC3595291
- DOI: 10.1371/journal.pone.0058608
Transcription factor-microRNA-target gene networks associated with ovarian cancer survival and recurrence
Abstract
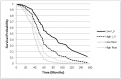
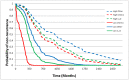
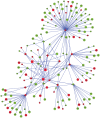
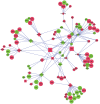
The identification of reliable transcriptome biomarkers requires the simultaneous consideration of regulatory and target elements including microRNAs (miRNAs), transcription factors (TFs), and target genes. A novel approach that integrates multivariate survival analysis, feature selection, and regulatory network visualization was used to identify reliable biomarkers of ovarian cancer survival and recurrence. Expression profiles of 799 miRNAs, 17,814 TFs and target genes and cohort clinical records on 272 patients diagnosed with ovarian cancer were simultaneously considered and results were validated on an independent group of 146 patients. Three miRNAs (hsa-miR-16, hsa-miR-22*, and ebv-miR-BHRF1-2*) were associated with both ovarian cancer survival and recurrence and 27 miRNAs were associated with either one hazard. Two miRNAs (hsa-miR-521 and hsa-miR-497) were cohort-dependent, while 28 were cohort-independent. This study confirmed 19 miRNAs previously associated with ovarian cancer and identified two miRNAs that have previously been associated with other cancer types. In total, the expression of 838 and 734 target genes and 12 and eight TFs were associated (FDR-adjusted P-value <0.05) with ovarian cancer survival and recurrence, respectively. Functional analysis highlighted the association between cellular and nucleotide metabolic processes and ovarian cancer. The more direct connections and higher centrality of the miRNAs, TFs and target genes in the survival network studied suggest that network-based approaches to prognosticate or predict ovarian cancer survival may be more effective than those for ovarian cancer recurrence. This study demonstrated the feasibility to infer reliable miRNA-TF-target gene networks associated with survival and recurrence of ovarian cancer based on the simultaneous analysis of co-expression profiles and consideration of the clinical characteristics of the patients.
Conflict of interest statement
Figures
References
-
- Hu X, Macdonald DM, Huettner PC, Feng Z, El Naqa IM, et al. (2009) A miR-200 microRNA cluster as prognostic marker in advanced ovarian cancer. Gynecol Oncol 114(3): 457–464. - PubMed
-
- Yang H, Kong W, He L, Zhao JJ, O’Donnell JD, et al. (2008) MicroRNA expression profiling in human ovarian cancer: miR-214 induces cell survival and cisplatin resistance by targeting PTEN. Cancer Res 68(2): 425–433. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Miscellaneous

