Complete chloroplast genome sequence of holoparasite Cistanche deserticola (Orobanchaceae) reveals gene loss and horizontal gene transfer from its host Haloxylon ammodendron (Chenopodiaceae)
- PMID: 23554920
- PMCID: PMC3598846
- DOI: 10.1371/journal.pone.0058747
Complete chloroplast genome sequence of holoparasite Cistanche deserticola (Orobanchaceae) reveals gene loss and horizontal gene transfer from its host Haloxylon ammodendron (Chenopodiaceae)
Abstract
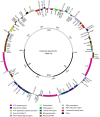
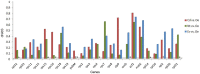
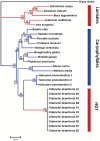
The central function of chloroplasts is to carry out photosynthesis, and its gene content and structure are highly conserved across land plants. Parasitic plants, which have reduced photosynthetic ability, suffer gene losses from the chloroplast (cp) genome accompanied by the relaxation of selective constraints. Compared with the rapid rise in the number of cp genome sequences of photosynthetic organisms, there are limited data sets from parasitic plants. PRINCIPAL FINDINGS/SIGNIFICANCE: Here we report the complete sequence of the cp genome of Cistanche deserticola, a holoparasitic desert species belonging to the family Orobanchaceae. The cp genome of C. deserticola is greatly reduced both in size (102,657 bp) and in gene content, indicating that all genes required for photosynthesis suffer from gene loss and pseudogenization, except for psbM. The striking difference from other holoparasitic plants is that it retains almost a full set of tRNA genes, and it has lower dN/dS for most genes than another close holoparasitic plant, E. virginiana, suggesting that Cistanche deserticola has undergone fewer losses, either due to a reduced level of holoparasitism, or to a recent switch to this life history. We also found that the rpoC2 gene was present in two copies within C. deserticola. Its own copy has much shortened and turned out to be a pseudogene. Another copy, which was not located in its cp genome, was a homolog of the host plant, Haloxylon ammodendron (Chenopodiaceae), suggesting that it was acquired from its host via a horizontal gene transfer.
Conflict of interest statement
Figures

Similar articles
-
Large-scale mRNA transfer between Haloxylon ammodendron (Chenopodiaceae) and herbaceous root holoparasite Cistanche deserticola (Orobanchaceae).iScience. 2022 Dec 27;26(1):105880. doi: 10.1016/j.isci.2022.105880. eCollection 2023 Jan 20. iScience. 2022. PMID: 36686392 Free PMC article.
-
The complete chloroplast genome of Caroxylon passerinum (Chenopodiaceae), an annual desert plant.Mitochondrial DNA B Resour. 2022 Feb 27;7(3):426-427. doi: 10.1080/23802359.2021.1994891. eCollection 2022. Mitochondrial DNA B Resour. 2022. PMID: 35252576 Free PMC article.
-
Combined Metabolome and Transcriptome Analysis Highlights the Host's Influence on Cistanche deserticola Metabolite Accumulation.Int J Mol Sci. 2023 Apr 27;24(9):7968. doi: 10.3390/ijms24097968. Int J Mol Sci. 2023. PMID: 37175675 Free PMC article.
-
Reductive evolution of chloroplasts in non-photosynthetic plants, algae and protists.Curr Genet. 2018 Apr;64(2):365-387. doi: 10.1007/s00294-017-0761-0. Epub 2017 Oct 12. Curr Genet. 2018. PMID: 29026976 Review.
-
Horizontal gene transfer from chloroplast to mitochondria of seagrasses in the yellow-Bohai seas.Genomics. 2024 Sep;116(5):110940. doi: 10.1016/j.ygeno.2024.110940. Epub 2024 Sep 18. Genomics. 2024. PMID: 39303860 Review.
Cited by
-
The plastid genome of mycoheterotrophic monocot Petrosavia stellaris exhibits both gene losses and multiple rearrangements.Genome Biol Evol. 2014 Jan;6(1):238-46. doi: 10.1093/gbe/evu001. Genome Biol Evol. 2014. PMID: 24398375 Free PMC article.
-
The Plastomes of Two Species in the Endoparasite Genus Pilostyles (Apodanthaceae) Each Retain Just Five or Six Possibly Functional Genes.Genome Biol Evol. 2015 Dec 12;8(1):189-201. doi: 10.1093/gbe/evv251. Genome Biol Evol. 2015. PMID: 26660355 Free PMC article.
-
Limited mitogenomic degradation in response to a parasitic lifestyle in Orobanchaceae.Sci Rep. 2016 Nov 3;6:36285. doi: 10.1038/srep36285. Sci Rep. 2016. PMID: 27808159 Free PMC article.
-
Convergent Plastome Evolution and Gene Loss in Holoparasitic Lennoaceae.Genome Biol Evol. 2018 Oct 1;10(10):2663-2670. doi: 10.1093/gbe/evy190. Genome Biol Evol. 2018. PMID: 30169817 Free PMC article.
-
Comparative and phylogenetic analyses of the chloroplast genomes of species of Paeoniaceae.Sci Rep. 2021 Jul 19;11(1):14643. doi: 10.1038/s41598-021-94137-0. Sci Rep. 2021. PMID: 34282194 Free PMC article.
References
-
- Raubeson LA, Jansen RK (2005) Chloroplast genomes of plants. In: Plant diversity and evolution: genotypic and phenotypic variation in higher plants. Henry RJ (Ed.). CABI Publishing, London.
-
- Krause K (2012) Plastid genomes of parasitic plants: a trail of reductions and losses. In: Organelle genetics. Bullerwell C (Ed.). Springer, Berlin.
-
- Jansen RK, Ruhlman TA (2012) Plastid Genomes of Seed Plants. In: Genomics of Chloroplasts and Mitochondria, Advances in Photosynthesis and Respiration. Bock R, Knoop V (Eds.). Springer, Berlin.
Publication types
MeSH terms
LinkOut - more resources
Full Text Sources
Other Literature Sources
Miscellaneous

