FlgM as a secretion moiety for the development of an inducible type III secretion system
- PMID: 23554966
- PMCID: PMC3595227
- DOI: 10.1371/journal.pone.0059034
FlgM as a secretion moiety for the development of an inducible type III secretion system
Abstract
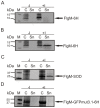
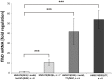
Regulation and assembly of the flagellar type III secretion system is one of the most investigated and best understood regulational cascades in molecular biology. Depending on the host organism, flagellar morphogenesis requires the interplay of more than 50 genes. Direct secretion of heterologous proteins to the supernatant is appealing due to protection against cellular proteases and simplified downstream processing. As Escherichia coli currently remains the predominant host organism used for recombinant prokaryotic protein expression, the generation of a strain that exhibits inducible flagellar secretion would be highly desirable for biotechnological applications. Here, we report the first engineered Escherichia coli mutant strain featuring flagellar morphogenesis upon addition of an external inducer. Using FlgM as a sensor for direct secretion in combination with this novel strain may represent a potent tool for significant improvements in future engineering of an inducible type III secretion for heterologous proteins.
Conflict of interest statement
Figures




Similar articles
-
Engineering the flagellar type III secretion system: improving capacity for secretion of recombinant protein.Microb Cell Fact. 2019 Jan 18;18(1):10. doi: 10.1186/s12934-019-1058-4. Microb Cell Fact. 2019. PMID: 30657054 Free PMC article.
-
Cellular levels and activity of the flagellar sigma factor FliA of Escherichia coli are controlled by FlgM-modulated proteolysis.Mol Microbiol. 2007 Jul;65(1):76-89. doi: 10.1111/j.1365-2958.2007.05770.x. Epub 2007 May 30. Mol Microbiol. 2007. PMID: 17537210
-
Analysis of factors that affect FlgM-dependent type III secretion for protein purification with Salmonella enterica serovar Typhimurium.J Bacteriol. 2014 Jul;196(13):2333-47. doi: 10.1128/JB.01572-14. Epub 2014 Apr 4. J Bacteriol. 2014. PMID: 24706743 Free PMC article.
-
Coupling of flagellar gene expression to flagellar assembly in Salmonella enterica serovar typhimurium and Escherichia coli.Microbiol Mol Biol Rev. 2000 Dec;64(4):694-708. doi: 10.1128/MMBR.64.4.694-708.2000. Microbiol Mol Biol Rev. 2000. PMID: 11104815 Free PMC article. Review.
-
Regulation of flagellar assembly.Curr Opin Microbiol. 2002 Apr;5(2):160-5. doi: 10.1016/s1369-5274(02)00302-8. Curr Opin Microbiol. 2002. PMID: 11934612 Review.
Cited by
-
Engineering the flagellar type III secretion system: improving capacity for secretion of recombinant protein.Microb Cell Fact. 2019 Jan 18;18(1):10. doi: 10.1186/s12934-019-1058-4. Microb Cell Fact. 2019. PMID: 30657054 Free PMC article.
-
Controlling tumor progression and recurrence in mice through combined treatment with a PD-L1 inhibitor and a designer Salmonella strain that delivers GM-CSF.Acta Pharm Sin B. 2024 Dec;14(12):5479-5492. doi: 10.1016/j.apsb.2024.07.011. Epub 2024 Jul 10. Acta Pharm Sin B. 2024. PMID: 39807328 Free PMC article.
-
Developing Gram-negative bacteria for the secretion of heterologous proteins.Microb Cell Fact. 2018 Dec 20;17(1):196. doi: 10.1186/s12934-018-1041-5. Microb Cell Fact. 2018. PMID: 30572895 Free PMC article. Review.
References
-
- Sperandio V, Torres AG, Kaper JB (2002) Quorum sensing Escherichia coli regulators B and C (QseBC): a novel two-component regulatory system involved in the regulation of flagella and motility by quorum sensing in E. coli. Mol Microbiol 43: 809–821. - PubMed
-
- Aldridge P, Hughes KT (2002) Regulation of flagellar assembly. Curr Opin Microbiol 5: 160–165. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials

