CD106 identifies a subpopulation of mesenchymal stem cells with unique immunomodulatory properties
- PMID: 23555021
- PMCID: PMC3595282
- DOI: 10.1371/journal.pone.0059354
CD106 identifies a subpopulation of mesenchymal stem cells with unique immunomodulatory properties
Abstract
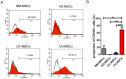
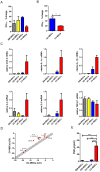
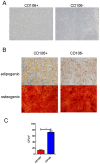
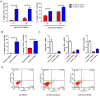
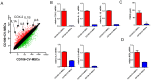
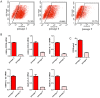
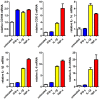
Mesenchymal stem cells (MSCs) reside in almost all of the body tissues, where they undergo self-renewal and multi-lineage differentiation. MSCs derived from different tissues share many similarities but also show some differences in term of biological properties. We aim to search for significant differences among various sources of MSCs and to explore their implications in physiopathology and clinical translation. We compared the phenotype and biological properties among different MSCs isolated from human term placental chorionic villi (CV), umbilical cord (UC), adult bone marrow (BM) and adipose (AD). We found that CD106 (VCAM-1) was expressed highest on the CV-MSCs, moderately on BM-MSCs, lightly on UC-MSCs and absent on AD-MSCs. CV-MSCs also showed unique immune-associated gene expression and immunomodulation. We thus separated CD106(+)cells and CD106(-)cells from CV-MSCs and compared their biological activities. Both two subpopulations were capable of osteogenic and adipogenic differentiation while CD106(+)CV-MSCs were more effective to modulate T helper subsets but possessed decreased colony formation capacity. In addition, CD106(+)CV-MSCs expressed more cytokines than CD106(-)CV-MSCs. These data demonstrate that CD106 identifies a subpopulation of CV-MSCs with unique immunoregulatory activity and reveal a previously unrecognized mechanism underlying immunomodulation of MSCs.
Conflict of interest statement
Figures
References
-
- Le Blanc K, Rasmusson I, Sundberg B, Gotherstrom C, Hassan M, et al. (2004) Treatment of severe acute graft-versus-host disease with third party haploidentical mesenchymal stem cells. Lancet 363: 1439–1441. - PubMed
-
- Zappia E, Casazza S, Pedemonte E, Benvenuto F, Bonanni I, et al. (2005) Mesenchymal stem cells ameliorate experimental autoimmune encephalomyelitis inducing T-cell anergy. Blood 106: 1755–1761. - PubMed
-
- Pittenger MF, Mackay AM, Beck SC, Jaiswal RK, Douglas R, et al. (1999) Multilineage potential of adult human mesenchymal stem cells. Science 284: 143–147. - PubMed
-
- Zuk PA, Zhu M, Mizuno H, Huang J, Futrell JW, et al. (2001) Multilineage cells from human adipose tissue: implications for cell-based therapies. Tissue Eng 7: 211–228. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Miscellaneous

