State-of-the-art fusion-finder algorithms sensitivity and specificity
- PMID: 23555082
- PMCID: PMC3595110
- DOI: 10.1155/2013/340620
State-of-the-art fusion-finder algorithms sensitivity and specificity
Abstract
Background: Gene fusions arising from chromosomal translocations have been implicated in cancer. RNA-seq has the potential to discover such rearrangements generating functional proteins (chimera/fusion). Recently, many methods for chimeras detection have been published. However, specificity and sensitivity of those tools were not extensively investigated in a comparative way.
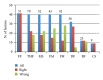
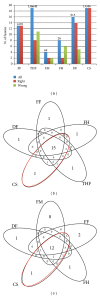
Results: We tested eight fusion-detection tools (FusionHunter, FusionMap, FusionFinder, MapSplice, deFuse, Bellerophontes, ChimeraScan, and TopHat-fusion) to detect fusion events using synthetic and real datasets encompassing chimeras. The comparison analysis run only on synthetic data could generate misleading results since we found no counterpart on real dataset. Furthermore, most tools report a very high number of false positive chimeras. In particular, the most sensitive tool, ChimeraScan, reports a large number of false positives that we were able to significantly reduce by devising and applying two filters to remove fusions not supported by fusion junction-spanning reads or encompassing large intronic regions.
Conclusions: The discordant results obtained using synthetic and real datasets suggest that synthetic datasets encompassing fusion events may not fully catch the complexity of RNA-seq experiment. Moreover, fusion detection tools are still limited in sensitivity or specificity; thus, there is space for further improvement in the fusion-finder algorithms.
Figures


References
-
- Mortazavi A, Williams BA, McCue K, Schaeffer L, Wold B. Mapping and quantifying mammalian transcriptomes by RNA-Seq. Nature Methods. 2008;5(7):621–628. - PubMed
-
- Abate F, Acquaviva A, Paciello G, et al. Bellerophontes: an RNA-Seq data analysis framework for chimeric transcripts discovery based on accurate fusion model. Bioinformatics. 2012;28(16):2114–2121. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources

