Folding pathways of a knotted protein with a realistic atomistic force field
- PMID: 23555232
- PMCID: PMC3605060
- DOI: 10.1371/journal.pcbi.1003002
Folding pathways of a knotted protein with a realistic atomistic force field
Abstract
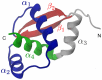
We report on atomistic simulation of the folding of a natively-knotted protein, MJ0366, based on a realistic force field. To the best of our knowledge this is the first reported effort where a realistic force field is used to investigate the folding pathways of a protein with complex native topology. By using the dominant-reaction pathway scheme we collected about 30 successful folding trajectories for the 82-amino acid long trefoil-knotted protein. Despite the dissimilarity of their initial unfolded configuration, these trajectories reach the natively-knotted state through a remarkably similar succession of steps. In particular it is found that knotting occurs essentially through a threading mechanism, involving the passage of the C-terminal through an open region created by the formation of the native [Formula: see text]-sheet at an earlier stage. The dominance of the knotting by threading mechanism is not observed in MJ0366 folding simulations using simplified, native-centric models. This points to a previously underappreciated role of concerted amino acid interactions, including non-native ones, in aiding the appropriate order of contact formation to achieve knotting.
Conflict of interest statement
The authors have declared that no competing interests exist.
Figures
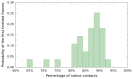
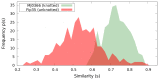
 distribution of DRP trajectories of the uknotted WW domain FIP35 .
distribution of DRP trajectories of the uknotted WW domain FIP35 .



 -sheet traps the
-sheet traps the  terminus on the “wrong” side of the
terminus on the “wrong” side of the  loop and prevents attaining the (native) knotted topology.
loop and prevents attaining the (native) knotted topology.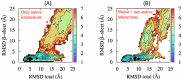
References
-
- Taylor WR (2007) Protein knots and fold complexity: some new twists. Comput Biol Chem 31: 151–162. - PubMed
-
- Mallam AL (2009) How does a knotted protein fold? FEBS J 276: 365–375. - PubMed
-
- Virnau P, Mallam A, Jackson S (2011) Structures and folding pathways of topologically knotted proteins. J Phys Condens Matter 23: 033101–033101. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources

