Genetic architecture of skin and eye color in an African-European admixed population
- PMID: 23555287
- PMCID: PMC3605137
- DOI: 10.1371/journal.pgen.1003372
Genetic architecture of skin and eye color in an African-European admixed population
Abstract
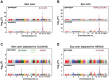
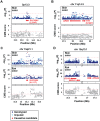
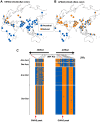
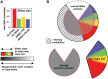
Variation in human skin and eye color is substantial and especially apparent in admixed populations, yet the underlying genetic architecture is poorly understood because most genome-wide studies are based on individuals of European ancestry. We study pigmentary variation in 699 individuals from Cape Verde, where extensive West African/European admixture has given rise to a broad range in trait values and genomic ancestry proportions. We develop and apply a new approach for measuring eye color, and identify two major loci (HERC2[OCA2] P = 2.3 × 10(-62), SLC24A5 P = 9.6 × 10(-9)) that account for both blue versus brown eye color and varying intensities of brown eye color. We identify four major loci (SLC24A5 P = 5.4 × 10(-27), TYR P = 1.1 × 10(-9), APBA2[OCA2] P = 1.5 × 10(-8), SLC45A2 P = 6 × 10(-9)) for skin color that together account for 35% of the total variance, but the genetic component with the largest effect (~44%) is average genomic ancestry. Our results suggest that adjacent cis-acting regulatory loci for OCA2 explain the relationship between skin and eye color, and point to an underlying genetic architecture in which several genes of moderate effect act together with many genes of small effect to explain ~70% of the estimated heritability.
Conflict of interest statement
The authors have declared that no competing interests exist.
Figures



References
-
- Jablonski NG, Chaplin G (2000) The evolution of human skin coloration. J Hum Evol 39: 57–106. - PubMed
-
- Parra EJ (2007) Human pigmentation variation: evolution, genetic basis, and implications for public health. Am J Phys Anthropol (Suppl 45) 85–105. - PubMed
-
- Di Rienzo A, Hudson RR (2005) An evolutionary framework for common diseases: the ancestral-susceptibility model. Trends Genet 21: 596–601. - PubMed
-
- Sulem P, Gudbjartsson DF, Stacey SN, Helgason A, Rafnar T, et al. (2007) Genetic determinants of hair, eye and skin pigmentation in Europeans. Nat Genet 39: 1443–1452. - PubMed
-
- Sulem P, Gudbjartsson DF, Stacey SN, Helgason A, Rafnar T, et al. (2008) Two newly identified genetic determinants of pigmentation in Europeans. Nat Genet 40: 835–837. - PubMed
Publication types
MeSH terms
Supplementary concepts
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

