Molecular networks of human muscle adaptation to exercise and age
- PMID: 23555298
- PMCID: PMC3605101
- DOI: 10.1371/journal.pgen.1003389
Molecular networks of human muscle adaptation to exercise and age
Erratum in
- PLoS Genet. 2013;9(4). doi: 10.1371/annotation/0dd3671e-1460-48fa-9d6a-2865dce78c07 doi: 10.1371/annotation/0dd3671e-1460-48fa-9d6a-2865dce78c07
Abstract
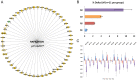
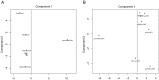
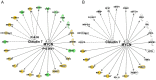
Physical activity and molecular ageing presumably interact to precipitate musculoskeletal decline in humans with age. Herein, we have delineated molecular networks for these two major components of sarcopenic risk using multiple independent clinical cohorts. We generated genome-wide transcript profiles from individuals (n = 44) who then undertook 20 weeks of supervised resistance-exercise training (RET). Expectedly, our subjects exhibited a marked range of hypertrophic responses (3% to +28%), and when applying Ingenuity Pathway Analysis (IPA) up-stream analysis to ~580 genes that co-varied with gain in lean mass, we identified rapamycin (mTOR) signaling associating with growth (P = 1.4 × 10(-30)). Paradoxically, those displaying most hypertrophy exhibited an inhibited mTOR activation signature, including the striking down-regulation of 70 rRNAs. Differential analysis found networks mimicking developmental processes (activated all-trans-retinoic acid (ATRA, Z-score = 4.5; P = 6 × 10(-13)) and inhibited aryl-hydrocarbon receptor signaling (AhR, Z-score = -2.3; P = 3 × 10(-7))) with RET. Intriguingly, as ATRA and AhR gene-sets were also a feature of endurance exercise training (EET), they appear to represent "generic" physical activity responsive gene-networks. For age, we found that differential gene-expression methods do not produce consistent molecular differences between young versus old individuals. Instead, utilizing two independent cohorts (n = 45 and n = 52), with a continuum of subject ages (18-78 y), the first reproducible set of age-related transcripts in human muscle was identified. This analysis identified ~500 genes highly enriched in post-transcriptional processes (P = 1 × 10(-6)) and with negligible links to the aforementioned generic exercise regulated gene-sets and some overlap with ribosomal genes. The RNA signatures from multiple compounds all targeting serotonin, DNA topoisomerase antagonism, and RXR activation were significantly related to the muscle age-related genes. Finally, a number of specific chromosomal loci, including 1q12 and 13q21, contributed by more than chance to the age-related gene list (P = 0.01-0.005), implying possible epigenetic events. We conclude that human muscle age-related molecular processes appear distinct from the processes regulated by those of physical activity.
Conflict of interest statement
The authors have declared that no competing interests exist.
Figures




References
-
- Gallagher IJ, Stephens NA, MacDonald AJ, Skipworth RJE, Husi H, et al. (2012) Suppression of skeletal muscle turnover in cancer cachexia: evidence from the transcriptome in sequential human muscle biopsies. Clinical cancer research: an official journal of the American Association for Cancer Research 18: 2817–2827. - PubMed
Publication types
MeSH terms
Substances
Associated data
- Actions
Grants and funding
- BB/X510697/1/BB_/Biotechnology and Biological Sciences Research Council/United Kingdom
- R01 HL047323/HL/NHLBI NIH HHS/United States
- HL-47317/HL/NHLBI NIH HHS/United States
- G1100015/MRC_/Medical Research Council/United Kingdom
- MR/K00414X/1/MRC_/Medical Research Council/United Kingdom
- HL-47323/HL/NHLBI NIH HHS/United States
- R01 HL047317/HL/NHLBI NIH HHS/United States
- MRC001873/MRC_/Medical Research Council/United Kingdom
- R01 HL047327/HL/NHLBI NIH HHS/United States
- HL-45670/HL/NHLBI NIH HHS/United States
- BB/C516779/1/BB_/Biotechnology and Biological Sciences Research Council/United Kingdom
- HL-47327/HL/NHLBI NIH HHS/United States
- R01 HL045670/HL/NHLBI NIH HHS/United States
- HL47321/HL/NHLBI NIH HHS/United States
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Molecular Biology Databases
Miscellaneous

