Differential evolutionary fate of an ancestral primate endogenous retrovirus envelope gene, the EnvV syncytin, captured for a function in placentation
- PMID: 23555306
- PMCID: PMC3610889
- DOI: 10.1371/journal.pgen.1003400
Differential evolutionary fate of an ancestral primate endogenous retrovirus envelope gene, the EnvV syncytin, captured for a function in placentation
Abstract
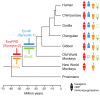
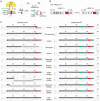
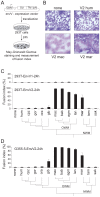
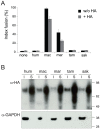
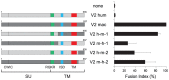
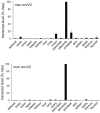
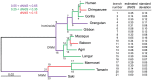
Syncytins are envelope genes of retroviral origin that have been co-opted for a role in placentation. They promote cell-cell fusion and are involved in the formation of a syncytium layer--the syncytiotrophoblast--at the materno-fetal interface. They were captured independently in eutherian mammals, and knockout mice demonstrated that they are absolutely required for placenta formation and embryo survival. Here we provide evidence that these "necessary" genes acquired "by chance" have a definite lifetime with diverse fates depending on the animal lineage, being both gained and lost in the course of evolution. Analysis of a retroviral envelope gene, the envV gene, present in primate genomes and belonging to the endogenous retrovirus type V (ERV-V) provirus, shows that this captured gene, which entered the primate lineage >45 million years ago, behaves as a syncytin in Old World monkeys, but lost its canonical fusogenic activity in other primate lineages, including humans. In the Old World monkeys, we show--by in situ analyses and ex vivo assays--that envV is both specifically expressed at the level of the placental syncytiotrophoblast and fusogenic, and that it further displays signs of purifying selection based on analysis of non-synonymous to synonymous substitution rates. We further show that purifying selection still operates in the primate lineages where the gene is no longer fusogenic, indicating that degeneracy of this ancestral syncytin is a slow, lineage-dependent, and multi-step process, in which the fusogenic activity would be the first canonical property of this retroviral envelope gene to be lost.
Conflict of interest statement
The authors have declared that no competing interests exist.
Figures


References
-
- Black SG, Arnaud F, Palmarini M, Spencer TE (2010) Endogenous retroviruses in trophoblast differentiation and placental development. Am J Reprod Immunol 64: 255–264 doi:10.1111/j.1600-0897.2010.00860.x. - DOI - PMC - PubMed
-
- Dupressoir A, Lavialle C, Heidmann T (2012) From ancestral infectious retroviruses to bona fide cellular genes: role of the captured syncytins in placentation. Placenta 33: 663–671 doi:10.1016/j.placenta.2012.05.005. - DOI - PubMed
-
- Mi S, Lee X, Li X, Veldman G, Finnerty H, et al. (2000) Syncytin is a captive retroviral envelope protein involved in human placental morphogenesis. Nature 17: 785–789 doi:10.1038/35001608. - DOI - PubMed
-
- Blond JL, Lavillette D, Cheynet V, Bouton O, Oriol G, et al. (2000) An envelope glycoprotein of the human endogenous retrovirus HERV-W is expressed in the human placenta and fuses cells expressing the type D mammalian retrovirus receptor. J Virol 74: 3321–3329 doi:10.1128/JVI.74.7.3321-3329.2000. - DOI - PMC - PubMed
-
- Mallet F, Bouton O, Prudhomme S, Cheynet V, Oriol G, et al. (2004) The endogenous retroviral locus ERVWE1 is a bona fide gene involved in hominoid placental physiology. Proc Natl Acad Sci U S A 101: 1731–1736 doi:10.1073/pnas.0305763101. - DOI - PMC - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

