Pseudomonas syringae pv. actinidiae from recent outbreaks of kiwifruit bacterial canker belong to different clones that originated in China
- PMID: 23555547
- PMCID: PMC3583860
- DOI: 10.1371/journal.pone.0057464
Pseudomonas syringae pv. actinidiae from recent outbreaks of kiwifruit bacterial canker belong to different clones that originated in China
Abstract
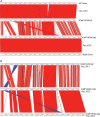
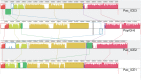
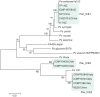
A recently emerged plant disease, bacterial canker of kiwifruit (Actinidia deliciosa and A. chinensis), is caused by Pseudomonas syringae pv. actinidiae (PSA). The disease was first reported in China and Japan in the 1980s. A severe outbreak of PSA began in Italy in 2008 and has spread to other European countries. PSA was found in both New Zealand and Chile in 2010. To study the evolution of the pathogen and analyse the transmission of PSA between countries, genomes of strains from China and Japan (where the genus Actinidia is endemic), Italy, New Zealand and Chile were sequenced. The genomes of PSA strains are very similar. However, all strains from New Zealand share several single nucleotide polymorphisms (SNPs) that distinguish them from all other PSA strains. Similarly, all the PSA strains from the 2008 Italian outbreak form a distinct clonal group and those from Chile form a third group. In addition to the rare SNPs present in the core genomes, there is abundant genetic diversity in a genomic island that is part of the accessory genome. The island from several Chinese strains is almost identical to the island present in the New Zealand strains. The island from a different Chinese strain is identical to the island present in the strains from the recent Italian outbreak. The Chilean strains of PSA carry a third variant of this island. These genomic islands are integrative conjugative elements (ICEs). Sequencing of these ICEs provides evidence of three recent horizontal transmissions of ICE from other strains of Pseudomonas syringae to PSA. The analyses of the core genome SNPs and the ICEs, combined with disease history, all support the hypothesis of an independent Chinese origin for both the Italian and the New Zealand outbreaks and suggest the Chilean strains also originate from China.
Conflict of interest statement
Figures




References
-
- Takikawa Y, Serizawa S, Ichikawa T (1989) Pseudomonas syringae pv. actinidiae pv. nov.: the causal bacterium of canker of kiwifruit in Japan. Annals of Phytopathological Society of Japan 55: 437–444.
-
- Wang Z, Tang X, Liu S (1992) Identification of the pathogenic bacterium for bacterial canker on Actinidia in Sichuan. Journal of Southwest Agricultural University: unpaginated.
-
- Anonymous (2011) Details on Pseudomonas syringae pv. actinidiae in China. EPPO Global Database article 2011/033. Available: http://gd3.eppo.int/reporting.php/article92. Accessed 11 September 2012.
-
- Koh JK, Cha BJ, Chung HJ, Lee DH (1994) Outbreak and spread of bacterial canker in kiwifruit. Korean Journal of Plant Pathology 10: 68–72.
Publication types
MeSH terms
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials
Miscellaneous

