A series of TA-based and zero-background vectors for plant functional genomics
- PMID: 23555713
- PMCID: PMC3612078
- DOI: 10.1371/journal.pone.0059576
A series of TA-based and zero-background vectors for plant functional genomics
Abstract
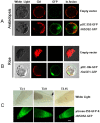
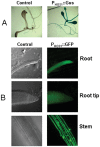
With the sequencing of genomes from many organisms now complete and the development of high-throughput sequencing, life science research has entered the functional post-genome era. Therefore, deciphering the function of genes and how they interact is in greater demand. To study an unknown gene, the basic methods are either overexpression or gene knockout by creating transgenic plants, and gene construction is usually the first step. Although traditional cloning techniques using restriction enzymes or a site-specific recombination system (Gateway or Clontech cloning technology) are highly useful for efficiently transferring DNA fragments into destination plasmids, the process is time consuming and expensive. To facilitate the procedure of gene construction, we designed a TA-based cloning system in which only one step was needed to subclone a DNA fragment into vectors. Such a cloning system was developed from the pGreen binary vector, which has a minimal size and facilitates construction manipulation, combined with the negative selection marker gene ccdB, which has the advantages of eliminating the self-ligation background and directly enabling high-efficiency TA cloning technology. We previously developed a set of transient and stable transformation vectors for constitutive gene expression, gene silencing, protein tagging, subcellular localization analysis and promoter activity detection. Our results show that such a system is highly efficient and serves as a high-throughput platform for transient or stable transformation in plants for functional genome research.
Conflict of interest statement
Figures





References
-
- Arabidopsis GI (2000) Analysis of the genome sequence of the flowering plant Arabidopsis thaliana. Nature 408: 796–815. - PubMed
-
- Marsischky G, LaBaer J (2004) Many paths to many clones: a comparative look at high-throughput cloning methods. Genome Res 14: 2020–2028. - PubMed
-
- Earley KW, Haag JR, Pontes O, Opper K, Juehne T, et al. (2006) Gateway-compatible vectors for plant functional genomics and proteomics. Plant J 45: 616–629. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources

