Impact of the 3D microenvironment on phenotype, gene expression, and EGFR inhibition of colorectal cancer cell lines
- PMID: 23555746
- PMCID: PMC3608563
- DOI: 10.1371/journal.pone.0059689
Impact of the 3D microenvironment on phenotype, gene expression, and EGFR inhibition of colorectal cancer cell lines
Abstract
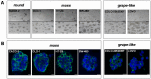
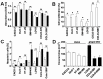
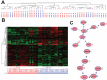
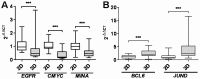
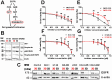
Three-dimensional (3D) tumor cell cultures grown in laminin-rich-extracellular matrix (lrECM) are considered to reflect human tumors more realistic as compared to cells grown as monolayer on plastic. Here, we systematically investigated the impact of ECM on phenotype, gene expression, EGFR signaling pathway, and on EGFR inhibition in commonly used colorectal cancer (CRC) cell lines. LrECM on-top (3D) culture assays were performed with the CRC cell lines SW-480, HT-29, DLD-1, LOVO, CACO-2, COLO-205 and COLO-206F. Morphology of lrECM cultivated CRC cell lines was determined by phase contrast and confocal laser scanning fluorescence microscopy. Proliferation of cells was examined by MTT assay, invasive capacity of the cell lines was assayed using Matrigel-coated Boyden chambers, and migratory activity was determined employing the Fence assay. Differential gene expression was analyzed at the transcriptional level by the Agilent array platform. EGFR was inhibited by using the specific small molecule inhibitor AG1478. A specific spheroid growth pattern was observed for all investigated CRC cell lines. DLD-1, HT-29 and SW-480 and CACO-2 exhibited a clear solid tumor cell formation, while LOVO, COLO-205 and COLO-206F were characterized by forming grape-like structures. Although the occurrence of a spheroid morphology did not correlate with an altered migratory, invasive, or proliferative capacity of CRC cell lines, gene expression was clearly altered in cells grown on lrECM as compared to 2D cultures. Interestingly, in KRAS wild-type cell lines, inhibition of EGFR was less effective in lrECM (3D) cultures as compared to 2D cell cultures. Thus, comparing both 2D and 3D cell culture models, our data support the influence of the ECM on cancer growth. Compared to conventional 2D cell culture, the lrECM (3D) cell culture model offers the opportunity to investigate permanent CRC cell lines under more physiological conditions, i.e. in the context of molecular therapeutic targets and their pharmacological inhibition.
Conflict of interest statement
Figures
References
-
- Masters JR (2000) Human cancer cell lines: fact and fantasy. Nat Rev Mol Cell Biol 1: 233–236. - PubMed
-
- Sharma SV, Haber DA, Settleman J (2010) Cell line-based platforms to evaluate the therapeutic efficacy of candidate anticancer agents. Nat Rev Cancer 10: 241–253. - PubMed
-
- Bellis SL (2004) Variant glycosylation: an underappreciated regulatory mechanism for beta1 integrins. Biochim Biophys Acta 1663: 52–60. - PubMed
-
- Benton G, George J, Kleinman HK, Arnaoutova IP (2009) Advancing science and technology via 3D culture on basement membrane matrix. J Cell Physiol 221: 18–25. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Molecular Biology Databases
Research Materials
Miscellaneous

