Deficient sumoylation of yeast 2-micron plasmid proteins Rep1 and Rep2 associated with their loss from the plasmid-partitioning locus and impaired plasmid inheritance
- PMID: 23555963
- PMCID: PMC3610928
- DOI: 10.1371/journal.pone.0060384
Deficient sumoylation of yeast 2-micron plasmid proteins Rep1 and Rep2 associated with their loss from the plasmid-partitioning locus and impaired plasmid inheritance
Abstract
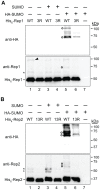
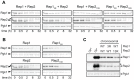
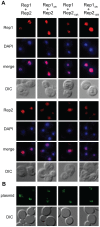
The 2-micron plasmid of the budding yeast Saccharomyces cerevisiae encodes copy-number amplification and partitioning systems that enable the plasmid to persist despite conferring no advantage to its host. Plasmid partitioning requires interaction of the plasmid Rep1 and Rep2 proteins with each other and with the plasmid-partitioning locus STB. Here we demonstrate that Rep1 stability is reduced in the absence of Rep2, and that both Rep proteins are sumoylated. Lysine-to-arginine substitutions in Rep1 and Rep2 that inhibited their sumoylation perturbed plasmid inheritance without affecting Rep protein stability or two-hybrid interaction between Rep1 and Rep2. One-hybrid and chromatin immunoprecipitation assays revealed that Rep1 was required for efficient retention of Rep2 at STB and that sumoylation-deficient mutants of Rep1 and Rep2 were impaired for association with STB. The normal co-localization of both Rep proteins with the punctate nuclear plasmid foci was also lost when Rep1 was sumoylation-deficient. The correlation of Rep protein sumoylation status with plasmid-partitioning locus association suggests a theme common to eukaryotic chromosome segregation proteins, sumoylated forms of which are found enriched at centromeres, and between the yeast 2-micron plasmid and viral episomes that depend on sumoylation of their maintenance proteins for persistence in their hosts.
Conflict of interest statement
Figures




Similar articles
-
The yeast 2-micron plasmid Rep2 protein has Rep1-independent partitioning function.Nucleic Acids Res. 2022 Oct 14;50(18):10571-10585. doi: 10.1093/nar/gkac810. Nucleic Acids Res. 2022. PMID: 36156142 Free PMC article.
-
The yeast 2-μm plasmid Raf protein contributes to plasmid inheritance by stabilizing the Rep1 and Rep2 partitioning proteins.Nucleic Acids Res. 2017 Oct 13;45(18):10518-10533. doi: 10.1093/nar/gkx703. Nucleic Acids Res. 2017. PMID: 29048592 Free PMC article.
-
The 2-μm plasmid encoded protein Raf1 regulates both stability and copy number of the plasmid by blocking the formation of the Rep1-Rep2 repressor complex.Nucleic Acids Res. 2017 Jul 7;45(12):7167-7179. doi: 10.1093/nar/gkx316. Nucleic Acids Res. 2017. PMID: 28472368 Free PMC article.
-
The 2 micron plasmid of Saccharomyces cerevisiae: a miniaturized selfish genome with optimized functional competence.Plasmid. 2013 Jul;70(1):2-17. doi: 10.1016/j.plasmid.2013.03.001. Epub 2013 Mar 27. Plasmid. 2013. PMID: 23541845 Review.
-
Insights into the DNA sequence elements required for partitioning and copy number control of the yeast 2-micron plasmid.Curr Genet. 2019 Aug;65(4):887-892. doi: 10.1007/s00294-019-00958-y. Epub 2019 Mar 26. Curr Genet. 2019. PMID: 30915516 Review.
Cited by
-
Genome maintenance in Saccharomyces cerevisiae: the role of SUMO and SUMO-targeted ubiquitin ligases.Nucleic Acids Res. 2017 Mar 17;45(5):2242-2261. doi: 10.1093/nar/gkw1369. Nucleic Acids Res. 2017. PMID: 28115630 Free PMC article. Review.
-
Evaluating Protein Extraction Techniques for Elucidating Proteomic Changes in Yeast Deletion Strains.Proteomes. 2025 Jul 1;13(3):28. doi: 10.3390/proteomes13030028. Proteomes. 2025. PMID: 40700272 Free PMC article.
-
The 2 micron plasmid: a selfish genetic element with an optimized survival strategy within Saccharomyces cerevisiae.Curr Genet. 2018 Feb;64(1):25-42. doi: 10.1007/s00294-017-0719-2. Epub 2017 Jun 8. Curr Genet. 2018. PMID: 28597305 Review.
-
The Ubiquitin Ligase (E3) Psh1p Is Required for Proper Segregation of both Centromeric and Two-Micron Plasmids in Saccharomyces cerevisiae.G3 (Bethesda). 2017 Nov 6;7(11):3731-3743. doi: 10.1534/g3.117.300227. G3 (Bethesda). 2017. PMID: 28928274 Free PMC article.
-
Replication-dependent and independent mechanisms for the chromosome-coupled persistence of a selfish genome.Nucleic Acids Res. 2016 Sep 30;44(17):8302-23. doi: 10.1093/nar/gkw694. Epub 2016 Aug 4. Nucleic Acids Res. 2016. PMID: 27492289 Free PMC article.
References
-
- Futcher AB (1988) The 2 micron circle plasmid of Saccharomyces cerevisiae . Yeast Chichester, England: 4: 27–40 10.1002/yea.320040104. - DOI - PubMed
-
- Broach JR, Volkert FC. (1991) Circular DNA plasmids of yeasts: Genome dynamics, protein synthesis and energetics. In: Broach JR, Pringle JR, Jones EW, editors. The molecular and cellular biology of the yeast Saccharomyces.Cold Spring Harbor, N.Y.: Cold Spring Harbor Laboratory press. pp. 297-331.
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials

