Generation of a suite of 3D computer-generated breast phantoms from a limited set of human subject data
- PMID: 23556929
- PMCID: PMC3625240
- DOI: 10.1118/1.4794924
Generation of a suite of 3D computer-generated breast phantoms from a limited set of human subject data
Abstract
Purpose: The authors previously reported on a three-dimensional computer-generated breast phantom, based on empirical human image data, including a realistic finite-element based compression model that was capable of simulating multimodality imaging data. The computerized breast phantoms are a hybrid of two phantom generation techniques, combining empirical breast CT (bCT) data with flexible computer graphics techniques. However, to date, these phantoms have been based on single human subjects. In this paper, the authors report on a new method to generate multiple phantoms, simulating additional subjects from the limited set of original dedicated breast CT data. The authors developed an image morphing technique to construct new phantoms by gradually transitioning between two human subject datasets, with the potential to generate hundreds of additional pseudoindependent phantoms from the limited bCT cases. The authors conducted a preliminary subjective assessment with a limited number of observers (n = 4) to illustrate how realistic the simulated images generated with the pseudoindependent phantoms appeared.
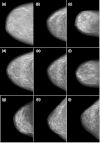
Methods: Several mesh-based geometric transformations were developed to generate distorted breast datasets from the original human subject data. Segmented bCT data from two different human subjects were used as the "base" and "target" for morphing. Several combinations of transformations were applied to morph between the "base' and "target" datasets such as changing the breast shape, rotating the glandular data, and changing the distribution of the glandular tissue. Following the morphing, regions of skin and fat were assigned to the morphed dataset in order to appropriately assign mechanical properties during the compression simulation. The resulting morphed breast was compressed using a finite element algorithm and simulated mammograms were generated using techniques described previously. Sixty-two simulated mammograms, generated from morphing three human subject datasets, were used in a preliminary observer evaluation where four board certified breast radiologists with varying amounts of experience ranked the level of realism (from 1 = "fake" to 10 = "real") of the simulated images.
Results: The morphing technique was able to successfully generate new and unique morphed datasets from the original human subject data. The radiologists evaluated the realism of simulated mammograms generated from the morphed and unmorphed human subject datasets and scored the realism with an average ranking of 5.87 ± 1.99, confirming that overall the phantom image datasets appeared more "real" than "fake." Moreover, there was not a significant difference (p > 0.1) between the realism of the unmorphed datasets (6.0 ± 1.95) compared to the morphed datasets (5.86 ± 1.99). Three of the four observers had overall average rankings of 6.89 ± 0.89, 6.9 ± 1.24, 6.76 ± 1.22, whereas the fourth observer ranked them noticeably lower at 2.94 ± 0.7.
Conclusions: This work presents a technique that can be used to generate a suite of realistic computerized breast phantoms from a limited number of human subjects. This suite of flexible breast phantoms can be used for multimodality imaging research to provide a known truth while concurrently producing realistic simulated imaging data.
Figures




References
-
- Berg W. A., Blume J. D., Cormack J. B., Mendelson E. B., Lehrer D., Bohm-Velez M., Pisano E. D., Jong R. A., Evans W. P., Morton M. J., Mahoney M. C., Hovanessian Larsen L., Barr R. G., Farria D. M., Marques H. S., and Boparai K., for the ACRIN 6666 Investigators, “Combined screening with ultrasound and mammography vs mammography alone in women at elevated risk of breast cancer,” J. Am. Med. Assoc. 299, 2151–2163 (2008). 10.1001/jama.299.18.2151 - DOI - PMC - PubMed
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical

