Invasion of E. coli biofilms by antibiotic resistance plasmids
- PMID: 23558148
- PMCID: PMC3687034
- DOI: 10.1016/j.plasmid.2013.03.003
Invasion of E. coli biofilms by antibiotic resistance plasmids
Abstract
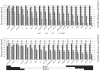
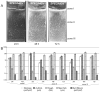
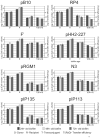
In spite of the contribution of plasmids to the spread of antibiotic resistance in human pathogens, little is known about the transferability of various drug resistance plasmids in bacterial biofilms. The goal of this study was to compare the efficiency of transfer of 19 multidrug resistance plasmids into Escherichia coli recipient biofilms and determine the effects of biofilm age, biofilm-donor exposure time, and donor-to-biofilm attachment on this process. An E. coli recipient biofilm was exposed separately to 19 E. coli donors, each with a different plasmid, and transconjugants were determined by plate counting. With few exceptions, plasmids that transferred well in a liquid environment also showed the highest transferability in biofilms. The difference in transfer frequency between the most and least transferable plasmid was almost a million-fold. The 'invasibility' of the biofilm by plasmids, or the proportion of biofilm cells that acquired plasmids within a few hours, depended not only on the type of plasmid, but also on the time of biofilm exposure to the donor and on the ability of the plasmid donor to attach to the biofilm, yet not on biofilm age. The efficiency of donor strain attachment to the biofilm was not affected by the presence of plasmids. The most invasive plasmid was pHH2-227, which based on genome sequence analysis is a hybrid between IncU-like and IncW plasmids. The wide range in transferability in an E. coli biofilm among plasmids needs to be taken into account in our fight against the spread of drug resistance.
Copyright © 2013 Elsevier Inc. All rights reserved.
Figures

References
-
- Andrup L. Conjugation in gram-positive bacteria and kinetics of plasmid transfer. APMIS Suppl. 1998;84:47–55. - PubMed
-
- Andrup L, Andersen K. A comparison of the kinetics of plasmid transfer in the conjugation systems encoded by the F plasmid from Escherichia coli and plasmid pCF10 from Enterococcus faecalis. Microbiology. 1999;145:2001–2009. - PubMed
-
- Babic A, et al. Direct visualization of horizontal gene transfer. Science. 2008;319:1533–1536. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Molecular Biology Databases

