Differential binding of the related transcription factors Pho4 and Cbf1 can tune the sensitivity of promoters to different levels of an induction signal
- PMID: 23558744
- PMCID: PMC3643608
- DOI: 10.1093/nar/gkt210
Differential binding of the related transcription factors Pho4 and Cbf1 can tune the sensitivity of promoters to different levels of an induction signal
Abstract
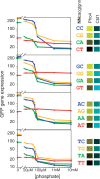
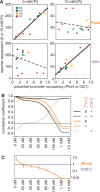
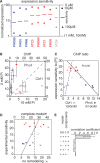
Transcription factors that belong to the same family typically have similar, but not identical, binding specificities. As such, they can be expected to compete differentially for binding to different variants of their binding sites. Pho4 is a yeast factor whose nuclear concentration is up-regulated in low phosphate, while the related factor, Cbf1, is constitutively expressed. We constructed 16 GFP-reporter genes containing all palindromic variants of the motif NNCACGTGNN, and determined their activities at a range of phosphate concentrations. Pho4 affinity did not explain expression data well except under fully induced conditions. However, reporter activity was quantitatively well explained under all conditions by a model in which Cbf1 itself has modest activating activity, and Pho4 and Cbf1 compete with one another. Chromatin immunoprecipitation and computational analyses of natural Pho4 target genes, along with the activities of the reporter constructs, indicates that genes differ in their sensitivity to intermediate induction signals in part because of differences in their affinity for Cbf1. The induction sensitivity of both natural Pho4 target genes and reporter genes was well explained only by a model that assumes a role for Cbf1 in remodeling chromatin. Our analyses highlight the importance of taking into account the activities of related transcription factors in explaining system-wide gene expression data.
Figures



References
-
- Johnson AD, Poteete AR, Lauer G, Sauer RT, Ackers GK, Ptashne M. λ Repressor and cro—components of an efficient molecular switch. Nature. 1981;294:217–223. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials

