The monomeric GIY-YIG homing endonuclease I-BmoI uses a molecular anchor and a flexible tether to sequentially nick DNA
- PMID: 23558745
- PMCID: PMC3664794
- DOI: 10.1093/nar/gkt186
The monomeric GIY-YIG homing endonuclease I-BmoI uses a molecular anchor and a flexible tether to sequentially nick DNA
Abstract
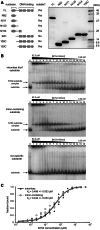
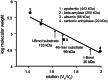
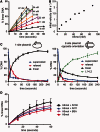
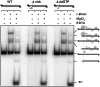
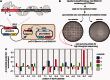
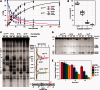
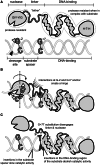
The GIY-YIG nuclease domain is found within protein scaffolds that participate in diverse cellular pathways and contains a single active site that hydrolyzes DNA by a one-metal ion mechanism. GIY-YIG homing endonucleases (GIY-HEs) are two-domain proteins with N-terminal GIY-YIG nuclease domains connected to C-terminal DNA-binding and they are thought to function as monomers. Using I-BmoI as a model GIY-HE, we test mechanisms by which the single active site is used to generate a double-strand break. We show that I-BmoI is partially disordered in the absence of substrate, and that the GIY-YIG domain alone has weak affinity for DNA. Significantly, we show that I-BmoI functions as a monomer at all steps of the reaction pathway and does not transiently dimerize or use sequential transesterification reactions to cleave substrate. Our results are consistent with the I-BmoI DNA-binding domain acting as a molecular anchor to tether the GIY-YIG domain to substrate, permitting rotation of the GIY-YIG domain to sequentially nick each DNA strand. These data highlight the mechanistic differences between monomeric GIY-HEs and dimeric or tetrameric GIY-YIG restriction enzymes, and they have implications for the use of the GIY-YIG domain in genome-editing applications.
Figures



References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

