LncRNA loc285194 is a p53-regulated tumor suppressor
- PMID: 23558749
- PMCID: PMC3643595
- DOI: 10.1093/nar/gkt182
LncRNA loc285194 is a p53-regulated tumor suppressor
Abstract
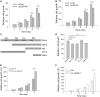
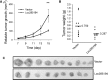
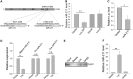
Protein-coding genes account for only a small part of the human genome, whereas the vast majority of transcripts make up the non-coding RNAs including long non-coding RNAs (lncRNAs). Accumulating evidence indicates that lncRNAs could play a critical role in regulation of cellular processes such as cell growth and apoptosis as well as cancer progression and metastasis. LncRNA loc285194 was previously shown to be within a tumor suppressor unit in osteosarcoma and to suppress tumor cell growth. However, it is unknown regarding the regulation of loc285194. Moreover, the underlying mechanism by which loc285194 functions as a potential tumor suppressor is elusive. In this study, we show that loc285194 is a p53 transcription target; ectopic expression of loc285194 inhibits tumor cell growth both in vitro and in vivo. Through deletion analysis, we identify an active region responsible for tumor cell growth inhibition within exon 4, which harbors two miR-211 binding sites. Importantly, this loc285194-mediated growth inhibition is in part due to specific suppression of miR-211. We further demonstrate a reciprocal repression between loc285194 and miR-211; in contrast to loc285194, miR-211 promotes cell growth. Finally, we detect downregulation of loc285194 in colon cancer specimens by quantitative PCR arrays and in situ hybridization of tissue microarrays. Together, these results suggest that loc285194 is a p53-regulated tumor suppressor, which acts in part through repression of miR-211.
Figures


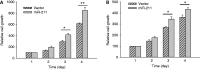

References
-
- Croce CM, Calin GA. miRNAs, cancer, and stem cell division. Cell. 2005;122:6–7. - PubMed
-
- Hammond SM. MicroRNAs as oncogenes. Curr. Opin. Genet. Dev. 2006;16:4–9. - PubMed
-
- Esquela-Kerscher A, Slack FJ. Oncomirs - microRNAs with a role in cancer. Nat. Rev. Cancer. 2006;6:259–269. - PubMed
-
- Gregory RI, Shiekhattar R. MicroRNA biogenesis and cancer. Cancer Res. 2005;65:3509–3512. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials
Miscellaneous

