Predicting the proteins of Angomonas deanei, Strigomonas culicis and their respective endosymbionts reveals new aspects of the trypanosomatidae family
- PMID: 23560078
- PMCID: PMC3616161
- DOI: 10.1371/journal.pone.0060209
Predicting the proteins of Angomonas deanei, Strigomonas culicis and their respective endosymbionts reveals new aspects of the trypanosomatidae family
Abstract
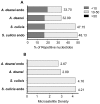
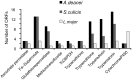
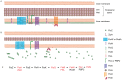
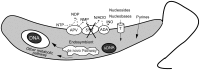
Endosymbiont-bearing trypanosomatids have been considered excellent models for the study of cell evolution because the host protozoan co-evolves with an intracellular bacterium in a mutualistic relationship. Such protozoa inhabit a single invertebrate host during their entire life cycle and exhibit special characteristics that group them in a particular phylogenetic cluster of the Trypanosomatidae family, thus classified as monoxenics. In an effort to better understand such symbiotic association, we used DNA pyrosequencing and a reference-guided assembly to generate reads that predicted 16,960 and 12,162 open reading frames (ORFs) in two symbiont-bearing trypanosomatids, Angomonas deanei (previously named as Crithidia deanei) and Strigomonas culicis (first known as Blastocrithidia culicis), respectively. Identification of each ORF was based primarily on TriTrypDB using tblastn, and each ORF was confirmed by employing getorf from EMBOSS and Newbler 2.6 when necessary. The monoxenic organisms revealed conserved housekeeping functions when compared to other trypanosomatids, especially compared with Leishmania major. However, major differences were found in ORFs corresponding to the cytoskeleton, the kinetoplast, and the paraflagellar structure. The monoxenic organisms also contain a large number of genes for cytosolic calpain-like and surface gp63 metalloproteases and a reduced number of compartmentalized cysteine proteases in comparison to other TriTryp organisms, reflecting adaptations to the presence of the symbiont. The assembled bacterial endosymbiont sequences exhibit a high A+T content with a total of 787 and 769 ORFs for the Angomonas deanei and Strigomonas culicis endosymbionts, respectively, and indicate that these organisms hold a common ancestor related to the Alcaligenaceae family. Importantly, both symbionts contain enzymes that complement essential host cell biosynthetic pathways, such as those for amino acid, lipid and purine/pyrimidine metabolism. These findings increase our understanding of the intricate symbiotic relationship between the bacterium and the trypanosomatid host and provide clues to better understand eukaryotic cell evolution.
Conflict of interest statement
Figures



References
-
- Wallace FG (1966) The trypanosomatid parasites of insects and arachnids. Experimental Parasitology 18: 124–193. - PubMed
-
- Teixeira MM, Borghesan TC, Ferreira RC, Santos MA, Takata CS, et al. (2011) Phylogenetic validation of the genera Angomonas and Strigomonas of trypanosomatids harboring bacterial endosymbionts with the description of new species of trypanosomatids and of proteobacterial symbionts. Protist 162: 503–524. - PubMed
-
- Edwards C, Chance B (1982) Evidence for the presence of two terminal oxidases in the trypanosomatid Crithidia oncopelti . Journal of General Microbiology 128: 1409–1414. - PubMed
-
- Fampa P, Correa-da-Silva MS, Lima DC, Oliveira SM, Motta MC, et al. (2003) Interaction of insect trypanosomatids with mosquitoes, sand fly and the respective insect cell lines. International Journal for Parasitology 33: 1019–1026. - PubMed
-
- de Azevedo-Martins AC, Frossard ML, de Souza W, Einicker-Lamas M, Motta MC (2007) Phosphatidylcholine synthesis in Crithidia deanei: the influence of the endosymbiont. FEMS Microbiology Letters 275: 229–236. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

