Phosphorylated C/EBPβ influences a complex network involving YY1 and USF2 in lung epithelial cells
- PMID: 23560079
- PMCID: PMC3613372
- DOI: 10.1371/journal.pone.0060211
Phosphorylated C/EBPβ influences a complex network involving YY1 and USF2 in lung epithelial cells
Abstract
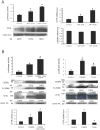
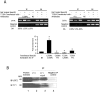
The promoter of the cystic fibrosis transmembrane conductance regulator gene CFTR is tightly controlled by regulators including CCAAT/enhancer binding proteins (C/EBPs). We previously reported that the transcription factors YY1 and USF2 affect CFTR expression. We can now demonstrate that C/EBPβ, a member of the CCAAT family, binds to the CFTR promoter and contributes to its transcriptional activity. Our data reveal that C/EBPβ cooperates with USF2 and acts antagonistically to YY1 in the control of CFTR expression. Interestingly, YY1, a strong repressor, fails to repress the CFTR activation induced by USF2 through DNA binding competition. Collectively, the data strongly suggest a model by which USF2 functionally interacts with YY1 blocking its inhibitory activity, in favour of C/EBPβ transactivation. Further investigation into the interactions between these three proteins revealed that phosphorylation of C/EBPβ influences the DNA occupancy of YY1 and favours the interaction between USF2 and YY1. This phosphorylation process has several implications in the CFTR transcriptional process, thus evoking an additional layer of complexity to the mechanisms influencing CFTR gene regulation.
Conflict of interest statement
Figures
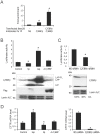

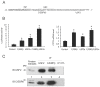
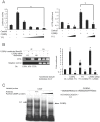
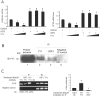

References
-
- Lekstrom-Himes J, Xanthopoulos KG (1998) Biological role of the CCAAT/enhancer-binding protein family of transcription factors. J Biol Chem 273: 28545–28548. - PubMed
-
- Cassel TN, Nord M (2003) C/EBP transcription factors in the lung epithelium. Am J Physiol Lung Cell Mol Physiol 285: L773–781. - PubMed
-
- Martis PC, Whitsett JA, Xu Y, Perl AK, Wan H, et al. (2006) C/EBPalpha is required for lung maturation at birth. Development 133: 1155–1164. - PubMed
-
- Pittman N, Shue G, LeLeiko NS, Walsh MJ (1995) Transcription of cystic fibrosis transmembrane conductance regulator requires a CCAAT-like element for both basal and cAMP-mediated regulation. J Biol Chem 270: 28848–28857. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources

