Mycobacterium tuberculosis ClpP proteases are co-transcribed but exhibit different substrate specificities
- PMID: 23560081
- PMCID: PMC3613350
- DOI: 10.1371/journal.pone.0060228
Mycobacterium tuberculosis ClpP proteases are co-transcribed but exhibit different substrate specificities
Abstract
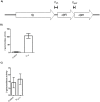
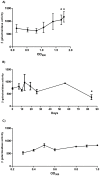
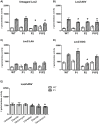
Caseinolytic (Clp) proteases are widespread energy-dependent proteases; the functional ATP-dependent protease is comprised of multimers of proteolytic and regulatory subunits. Mycobacterium tuberculosis has two ClpP proteolytic subunits (ClpP1 and ClpP2), with both being essential for growth in vitro. ClpP1 and clpP2 are arranged in an apparent operon; we demonstrated that the two genes are co-expressed under normal growth conditions. We identified a single promoter region for the clpP1P2 operon; no promoter was detected upstream of clpP2 demonstrating that independent expression of clpP1 and clpP2 was highly unlikely. Promoter activity was not induced by heat shock or oxidative stress. We identified a regulatory region upstream of the promoter with a consensus sequence matching the ClgR regulator motif; we determined the limits of the region by mutagenesis and confirmed that positive regulation of the promoter occurs in M. tuberculosis. We developed a reporter system to monitor ClpP1 and ClpP2 enzymatic activities based on LacZ incorporating ssrAtag sequences. We showed that whilst both ClpP1 and ClpP2 degrade SsrA-tagged LacZ, ClpP2 (but not ClpP1) degrades untagged proteins. Our data suggest that the two proteolytic subunits display different substrate specificities and therefore have different, but overlapping roles in M. tuberculosis.
Conflict of interest statement
Figures




Similar articles
-
The Mycobacterium tuberculosis ClpP1P2 Protease Interacts Asymmetrically with Its ATPase Partners ClpX and ClpC1.PLoS One. 2015 May 1;10(5):e0125345. doi: 10.1371/journal.pone.0125345. eCollection 2015. PLoS One. 2015. PMID: 25933022 Free PMC article.
-
Assembly and proteolytic processing of mycobacterial ClpP1 and ClpP2.BMC Biochem. 2011 Dec 1;12:61. doi: 10.1186/1471-2091-12-61. BMC Biochem. 2011. PMID: 22132756 Free PMC article.
-
ClgR, a novel regulator of clp and lon expression in Streptomyces.J Bacteriol. 2004 May;186(10):3238-48. doi: 10.1128/JB.186.10.3238-3248.2004. J Bacteriol. 2004. PMID: 15126487 Free PMC article.
-
Mycobacterium tuberculosis ClpP1 and ClpP2 function together in protein degradation and are required for viability in vitro and during infection.PLoS Pathog. 2012 Feb;8(2):e1002511. doi: 10.1371/journal.ppat.1002511. Epub 2012 Feb 16. PLoS Pathog. 2012. PMID: 22359499 Free PMC article.
-
Long range transcriptional control of virulence critical genes in Mycobacterium tuberculosis by nucleoid-associated proteins?Virulence. 2012 Jul 1;3(4):408-10. doi: 10.4161/viru.20918. Epub 2012 Jun 22. Virulence. 2012. PMID: 22722242 Free PMC article. Review. No abstract available.
Cited by
-
The Mycobacterium tuberculosis ClpP1P2 Protease Interacts Asymmetrically with Its ATPase Partners ClpX and ClpC1.PLoS One. 2015 May 1;10(5):e0125345. doi: 10.1371/journal.pone.0125345. eCollection 2015. PLoS One. 2015. PMID: 25933022 Free PMC article.
-
AAA+ Machines of Protein Destruction in Mycobacteria.Front Mol Biosci. 2017 Jul 19;4:49. doi: 10.3389/fmolb.2017.00049. eCollection 2017. Front Mol Biosci. 2017. PMID: 28770209 Free PMC article. Review.
-
Clostridium difficile ClpP Homologues are Capable of Uncoupled Activity and Exhibit Different Levels of Susceptibility to Acyldepsipeptide Modulation.ACS Infect Dis. 2019 Jan 11;5(1):79-89. doi: 10.1021/acsinfecdis.8b00199. Epub 2018 Nov 26. ACS Infect Dis. 2019. PMID: 30411608 Free PMC article.
-
Targeting the Proteostasis Network for Mycobacterial Drug Discovery.ACS Infect Dis. 2018 Apr 13;4(4):478-498. doi: 10.1021/acsinfecdis.7b00231. Epub 2018 Mar 2. ACS Infect Dis. 2018. PMID: 29465983 Free PMC article. Review.
-
Recent Insights into the Structure and Function of Mycobacterial Membrane Proteins Facilitated by Cryo-EM.J Membr Biol. 2021 Jun;254(3):321-341. doi: 10.1007/s00232-021-00179-w. Epub 2021 May 5. J Membr Biol. 2021. PMID: 33954837 Free PMC article. Review.
References
-
- WHO (2011) Global tuberculosis control. Available at: http://www.who.int/tb/publications/global_report/en/index.html (Accessed 1st August 2012).
-
- Porankiewicz J, Wang J, Clarke AK (1999) New insights into the ATP-dependent Clp protease: Escherichia coli and beyond. Mol Microbiol 32: 449–458. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Miscellaneous

