Quantitative design of regulatory elements based on high-precision strength prediction using artificial neural network
- PMID: 23560087
- PMCID: PMC3613377
- DOI: 10.1371/journal.pone.0060288
Quantitative design of regulatory elements based on high-precision strength prediction using artificial neural network
Abstract
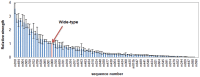
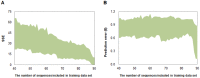
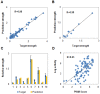
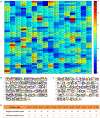
Accurate and controllable regulatory elements such as promoters and ribosome binding sites (RBSs) are indispensable tools to quantitatively regulate gene expression for rational pathway engineering. Therefore, de novo designing regulatory elements is brought back to the forefront of synthetic biology research. Here we developed a quantitative design method for regulatory elements based on strength prediction using artificial neural network (ANN). One hundred mutated Trc promoter & RBS sequences, which were finely characterized with a strength distribution from 0 to 3.559 (relative to the strength of the original sequence which was defined as 1), were used for model training and test. A precise strength prediction model, NET90_19_576, was finally constructed with high regression correlation coefficients of 0.98 for both model training and test. Sixteen artificial elements were in silico designed using this model. All of them were proved to have good consistency between the measured strength and our desired strength. The functional reliability of the designed elements was validated in two different genetic contexts. The designed parts were successfully utilized to improve the expression of BmK1 peptide toxin and fine-tune deoxy-xylulose phosphate pathway in Escherichia coli. Our results demonstrate that the methodology based on ANN model can de novo and quantitatively design regulatory elements with desired strengths, which are of great importance for synthetic biology applications.
Conflict of interest statement
Figures

Similar articles
-
Significant expression of a Chinese scorpion peptide, BmK1, in Escherichia coli through promoter engineering and gene dosage strategy.Biotechnol Appl Biochem. 2014 Jul-Aug;61(4):466-73. doi: 10.1002/bab.1194. Epub 2014 Mar 25. Biotechnol Appl Biochem. 2014. PMID: 24372571 Free PMC article.
-
Promoter knock-in: a novel rational method for the fine tuning of genes.BMC Biotechnol. 2010 Mar 24;10:26. doi: 10.1186/1472-6750-10-26. BMC Biotechnol. 2010. PMID: 20334648 Free PMC article.
-
Fine-Tuning Multi-Gene Clusters via Well-Characterized Gene Expression Regulatory Elements: Case Study of the Arginine Synthesis Pathway in C. glutamicum.ACS Synth Biol. 2021 Jan 15;10(1):38-48. doi: 10.1021/acssynbio.0c00405. Epub 2020 Dec 31. ACS Synth Biol. 2021. PMID: 33382575
-
Recent advances in the applications of promoter engineering for the optimization of metabolite biosynthesis.World J Microbiol Biotechnol. 2019 Jan 31;35(2):33. doi: 10.1007/s11274-019-2606-0. World J Microbiol Biotechnol. 2019. PMID: 30706208 Review.
-
Consensus architecture of promoters and transcription units in Escherichia coli: design principles for synthetic biology.Mol Biosyst. 2017 Mar 28;13(4):665-676. doi: 10.1039/c6mb00789a. Mol Biosyst. 2017. PMID: 28256660 Review.
Cited by
-
Advances and computational tools towards predictable design in biological engineering.Comput Math Methods Med. 2014;2014:369681. doi: 10.1155/2014/369681. Epub 2014 Aug 3. Comput Math Methods Med. 2014. PMID: 25161694 Free PMC article. Review.
-
Synthetic promoter design for new microbial chassis.Biochem Soc Trans. 2016 Jun 15;44(3):731-7. doi: 10.1042/BST20160042. Biochem Soc Trans. 2016. PMID: 27284035 Free PMC article. Review.
-
Synthetic promoter design in Escherichia coli based on a deep generative network.Nucleic Acids Res. 2020 Jul 9;48(12):6403-6412. doi: 10.1093/nar/gkaa325. Nucleic Acids Res. 2020. PMID: 32424410 Free PMC article.
-
Synergistic Synthetic Biology: Units in Concert.Front Bioeng Biotechnol. 2013 Oct 16;1:11. doi: 10.3389/fbioe.2013.00011. eCollection 2013. Front Bioeng Biotechnol. 2013. PMID: 25022769 Free PMC article. Review.
-
Dynamic modulation of enzyme activity by synthetic CRISPR-Cas6 endonucleases.Nat Chem Biol. 2022 May;18(5):492-500. doi: 10.1038/s41589-022-01005-7. Epub 2022 Apr 25. Nat Chem Biol. 2022. PMID: 35468950
References
-
- Dehli T, Solem C, Jensen PR (2012) Tunable promoters in synthetic and systems biology. Subcell Biochem 64: 181–201. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Miscellaneous

