The evolution of an osmotically inducible dps in the genus Streptomyces
- PMID: 23560105
- PMCID: PMC3613396
- DOI: 10.1371/journal.pone.0060772
The evolution of an osmotically inducible dps in the genus Streptomyces
Abstract
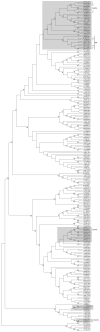
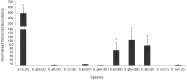
Dps proteins are found almost ubiquitously in bacterial genomes and there is now an appreciation of their multifaceted roles in various stress responses. Previous studies have shown that this family of proteins assemble into dodecamers and their quaternary structure is entirely critical to their function. Moreover, the numbers of dps genes per bacterial genome is variable; even amongst closely related species - however, for many genera this enigma is yet to be satisfactorily explained. We reconstruct the most probable evolutionary history of Dps in Streptomyces genomes. Typically, these bacteria encode for more than one Dps protein. We offer the explanation that variation in the number of dps per genome among closely related Streptomyces can be explained by gene duplication or lateral acquisition, and the former preceded a subsequent shift in expression patterns for one of the resultant paralogs. We show that the genome of S. coelicolor encodes for three Dps proteins including a tailless Dps. Our in vivo observations show that the tailless protein, unlike the other two Dps in S. coelicolor, does not readily oligomerise. Phylogenetic and bioinformatic analyses combined with expression studies indicate that in several Streptomyces species at least one Dps is significantly over-expressed during osmotic shock, but the identity of the ortholog varies. In silico analysis of dps promoter regions coupled with gene expression studies of duplicated dps genes shows that paralogous gene pairs are expressed differentially and this correlates with the presence of a sigB promoter. Lastly, we identify a rare novel clade of Dps and show that a representative of these proteins in S. coelicolor possesses a dodecameric quaternary structure of high stability.
Conflict of interest statement
Figures




References
-
- Matin A (1991) The molecular basis of carbon-starvation-induced general resistance in Escherichia coli. Molecular Microbiology 5: 3–10. - PubMed
-
- Almirón M, Link AJ, Furlong D, Kolter R (1992) A novel DNA-binding protein with regulatory and protective roles in starved Escherichia coli. Genes & Development 6: 2646–2654. - PubMed
-
- Chowdhury RP, Saraswathi R, Chatterji D (2010) Mycobacterial stress regulation: The Dps "twin sister" defense mechanism and structure-function relationship. IUBMB Life 62: 67–77. - PubMed
-
- Calhoun LN, Kwon YM (2010) Structure, function and regulation of the DNA-binding protein Dps and its role in acid and oxidative stress resistance in Escherichia coli: a review. J Appl Microbiol 110: 375–386. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Miscellaneous

