GIGI: an approach to effective imputation of dense genotypes on large pedigrees
- PMID: 23561844
- PMCID: PMC3617386
- DOI: 10.1016/j.ajhg.2013.02.011
GIGI: an approach to effective imputation of dense genotypes on large pedigrees
Abstract
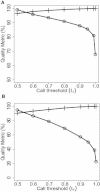
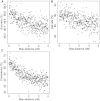
Recent emergence of the common-disease-rare-variant hypothesis has renewed interest in the use of large pedigrees for identifying rare causal variants. Genotyping with modern sequencing platforms is increasingly common in the search for such variants but remains expensive and often is limited to only a few subjects per pedigree. In population-based samples, genotype imputation is widely used so that additional genotyping is not needed. We now introduce an analogous approach that enables computationally efficient imputation in large pedigrees. Our approach samples inheritance vectors (IVs) from a Markov Chain Monte Carlo sampler by conditioning on genotypes from a sparse set of framework markers. Missing genotypes are probabilistically inferred from these IVs along with observed dense genotypes that are available on a subset of subjects. We implemented our approach in the Genotype Imputation Given Inheritance (GIGI) program and evaluated the approach on both simulated and real large pedigrees. With a real pedigree, we also compared imputed results obtained from this approach with those from the population-based imputation program BEAGLE. We demonstrated that our pedigree-based approach imputes many alleles with high accuracy. It is much more accurate for calling rare alleles than is population-based imputation and does not require an outside reference sample. We also evaluated the effect of varying other parameters, including the marker type and density of the framework panel, threshold for calling genotypes, and population allele frequencies. By leveraging information from existing genotypes already assayed on large pedigrees, our approach can facilitate cost-effective use of sequence data in the pursuit of rare causal variants.
Copyright © 2013 The American Society of Human Genetics. Published by Elsevier Inc. All rights reserved.
Figures



Similar articles
-
Whole-genome characterization in pedigreed non-human primates using genotyping-by-sequencing (GBS) and imputation.BMC Genomics. 2016 Aug 24;17(1):676. doi: 10.1186/s12864-016-2966-x. BMC Genomics. 2016. PMID: 27558348 Free PMC article.
-
A statistical framework to guide sequencing choices in pedigrees.Am J Hum Genet. 2014 Feb 6;94(2):257-67. doi: 10.1016/j.ajhg.2014.01.005. Am J Hum Genet. 2014. PMID: 24507777 Free PMC article.
-
Detection of Mendelian consistent genotyping errors in pedigrees.Genet Epidemiol. 2014 May;38(4):291-9. doi: 10.1002/gepi.21806. Epub 2014 Apr 9. Genet Epidemiol. 2014. PMID: 24718985 Free PMC article.
-
Two-stage strategy using denoising autoencoders for robust reference-free genotype imputation with missing input genotypes.J Hum Genet. 2024 Oct;69(10):511-518. doi: 10.1038/s10038-024-01261-6. Epub 2024 Jun 25. J Hum Genet. 2024. PMID: 38918526 Free PMC article. Review.
-
Linkage analysis with sequential imputation.Genet Epidemiol. 2003 Jul;25(1):25-35. doi: 10.1002/gepi.10249. Genet Epidemiol. 2003. PMID: 12813724 Review.
Cited by
-
Accurate Genotype Imputation in Multiparental Populations from Low-Coverage Sequence.Genetics. 2018 Sep;210(1):71-82. doi: 10.1534/genetics.118.300885. Epub 2018 Jul 25. Genetics. 2018. PMID: 30045858 Free PMC article.
-
Whole-genome characterization in pedigreed non-human primates using genotyping-by-sequencing (GBS) and imputation.BMC Genomics. 2016 Aug 24;17(1):676. doi: 10.1186/s12864-016-2966-x. BMC Genomics. 2016. PMID: 27558348 Free PMC article.
-
PBAP: a pipeline for file processing and quality control of pedigree data with dense genetic markers.Bioinformatics. 2015 Dec 1;31(23):3790-8. doi: 10.1093/bioinformatics/btv444. Epub 2015 Jul 30. Bioinformatics. 2015. PMID: 26231429 Free PMC article.
-
Genomic Exploration of Essential Hypertension in African-Brazilian Quilombo Populations: A Comprehensive Approach with Pedigree Analysis and Family-Based Association Studies.medRxiv [Preprint]. 2025 Feb 4:2024.06.26.24309531. doi: 10.1101/2024.06.26.24309531. medRxiv. 2025. Update in: J Am Heart Assoc. 2025 Apr;14(7):e036193. doi: 10.1161/JAHA.124.036193. PMID: 38978678 Free PMC article. Updated. Preprint.
-
Contribution of common and rare variants to bipolar disorder susceptibility in extended pedigrees from population isolates.Transl Psychiatry. 2020 Feb 24;10(1):74. doi: 10.1038/s41398-020-0758-1. Transl Psychiatry. 2020. PMID: 32094344 Free PMC article.
References
-
- Amberger J., Bocchini C., Hamosh A. A new face and new challenges for Online Mendelian Inheritance in Man (OMIM®) Hum. Mutat. 2011;32:564–567. - PubMed
-
- Collins F.S., Guyer M.S., Charkravarti A. Variations on a theme: cataloging human DNA sequence variation. Science. 1997;278:1580–1581. - PubMed
-
- Cohen J.C., Kiss R.S., Pertsemlidis A., Marcel Y.L., McPherson R., Hobbs H.H. Multiple rare alleles contribute to low plasma levels of HDL cholesterol. Science. 2004;305:869–872. - PubMed
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

