An evolutionary shift in the regulation of the Hippo pathway between mice and flies
- PMID: 23563179
- PMCID: PMC4613760
- DOI: 10.1038/onc.2013.82
An evolutionary shift in the regulation of the Hippo pathway between mice and flies
Abstract
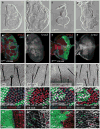
The Hippo pathway plays a key role in controlling organ growth in many animal species and its deregulation is associated with different types of cancer. Understanding the regulation of the Hippo pathway and discovering upstream regulators is thus a major quest. Interestingly, while the core of the Hippo pathway contains a highly conserved kinase cascade, different components have been identified as upstream regulators in Drosophila and vertebrates. However, whether the regulation of the Hippo pathway is indeed different between Drosophila and vertebrates or whether these differences are due to our limited analysis of these components in different organisms is not known. Here we show that the mouse Fat4 cadherin, the ortholog of the Hippo pathway regulator Fat in Drosophila, does not apparently regulate the Hippo pathway in the murine liver. In fact, we uncovered an evolutionary shift in many of the known upstream regulators at the base of the arthropod lineage. In this evolutionary transition, Fat and the adaptor protein Expanded gained novel domains that connected them to the Hippo pathway, whereas the cell-adhesion receptor Echinoid evolved as a new protein. Subsequently, the junctional adaptor protein Angiomotin (Amot) was lost and the downstream effector Yap lost its PDZ-binding motif that interacts with cell junction proteins. We conclude that fundamental differences exist in the upstream regulatory mechanisms of Hippo signaling between Drosophila and vertebrates.
Conflict of interest statement
Figures



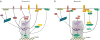
References
-
- Genevet A, Tapon N. The Hippo pathway and apico-basal cell polarity. Biochem J. 2011;436:213–224. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

