p190-B RhoGAP regulates the functional composition of the mesenchymal microenvironment
- PMID: 23563238
- PMCID: PMC3919554
- DOI: 10.1038/leu.2013.103
p190-B RhoGAP regulates the functional composition of the mesenchymal microenvironment
Abstract
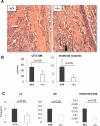
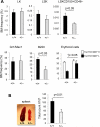
Hematopoiesis is regulated by components of the microenvironment, so-called niche. Here, we show that p190-B GTPase-activating protein (p190-B) deletion in mice causes hematopoietic failure during ontogeny, in p190-B(-/-) fetal liver and bones, and in p190-B(+/-) adult bones and spleen. These defects are non-cell autonomous, as we previously showed that transplantation of p190-B(-/-) hematopoietic cells into wild-type (WT) hosts leads to normal hematopoiesis. Coculture of mesenchymal stem (MSC)/progenitor cells and wild-type bone marrow (BM) cells reveals that p190-B(-/-) MSCs are dysfunctional in supporting hematopoiesis owing to impaired Wnt signaling. Furthermore, p190-B loss causes alteration in BM niche composition, including abnormal colony-forming unit (CFU)-fibroblast, CFU-adipocyte and CFU-osteoblast numbers. This is due to altered MSC lineage fate specification to osteoblast and adipocyte lineages. Thus, p190-B organizes a functional mesenchymal/microenvironment for normal hematopoiesis during development.
Figures
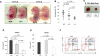


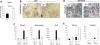

Similar articles
-
Lack of Adipocytes Alters Hematopoiesis in Lipodystrophic Mice.Front Immunol. 2018 Nov 13;9:2573. doi: 10.3389/fimmu.2018.02573. eCollection 2018. Front Immunol. 2018. PMID: 30483254 Free PMC article.
-
TGF-β Signaling Plays an Essential Role in the Lineage Specification of Mesenchymal Stem/Progenitor Cells in Fetal Bone Marrow.Stem Cell Reports. 2019 Jul 9;13(1):48-60. doi: 10.1016/j.stemcr.2019.05.017. Epub 2019 Jun 13. Stem Cell Reports. 2019. PMID: 31204302 Free PMC article.
-
Impact of interactions of cellular components of the bone marrow microenvironment on hematopoietic stem and progenitor cell function.Blood. 2010 Apr 22;115(16):3239-48. doi: 10.1182/blood-2009-09-246173. Epub 2010 Feb 12. Blood. 2010. PMID: 20154218 Free PMC article.
-
Regulation of hematopoiesis in endosteal microenvironments.Int J Hematol. 2014 Jun;99(6):679-84. doi: 10.1007/s12185-014-1583-1. Epub 2014 Apr 24. Int J Hematol. 2014. PMID: 24760425 Review.
-
From Marrow to Bone and Fat: Exploring the Multifaceted Roles of Leptin Receptor Positive Bone Marrow Mesenchymal Stromal Cells.Cells. 2024 May 24;13(11):910. doi: 10.3390/cells13110910. Cells. 2024. PMID: 38891042 Free PMC article. Review.
Cited by
-
Rho GTPase Regulators and Effectors in Autism Spectrum Disorders: Animal Models and Insights for Therapeutics.Cells. 2020 Mar 31;9(4):835. doi: 10.3390/cells9040835. Cells. 2020. PMID: 32244264 Free PMC article. Review.
-
CD147 promotes cell motility via upregulation of p190-B RhoGAP in hepatocellular carcinoma.Cancer Cell Int. 2016 Sep 6;16(1):69. doi: 10.1186/s12935-016-0344-z. eCollection 2016. Cancer Cell Int. 2016. PMID: 27601938 Free PMC article.
-
ARAP3 functions in hematopoietic stem cells.PLoS One. 2014 Dec 26;9(12):e116107. doi: 10.1371/journal.pone.0116107. eCollection 2014. PLoS One. 2014. PMID: 25542002 Free PMC article.
-
HES1 is required for mouse fetal hematopoiesis.Stem Cell Res Ther. 2024 Jul 29;15(1):235. doi: 10.1186/s13287-024-03836-8. Stem Cell Res Ther. 2024. PMID: 39075526 Free PMC article.
-
p190-B RhoGAP and intracellular cytokine signals balance hematopoietic stem and progenitor cell self-renewal and differentiation.Nat Commun. 2017 Feb 8;8:14382. doi: 10.1038/ncomms14382. Nat Commun. 2017. PMID: 28176763 Free PMC article.
References
-
- Kiel MJ, Morrison SJ. Uncertainty in the niches that maintain haematopoietic stem cells. Nat Rev Immunol. 2008;8(4):290–301. - PubMed
-
- Mikkola HK, Orkin SH. The journey of developing hematopoietic stem cells. Development. 2006;133(19):3733–44. - PubMed
-
- Caplan AI. Mesenchymal stem cells. J Orthop Res. 1991;9(5):641–50. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Molecular Biology Databases

