Genetic architecture of olfactory behavior in Drosophila melanogaster: differences and similarities across development
- PMID: 23563598
- PMCID: PMC3691330
- DOI: 10.1007/s10519-013-9592-z
Genetic architecture of olfactory behavior in Drosophila melanogaster: differences and similarities across development
Abstract
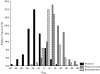
In the holometabolous insect Drosophila melanogaster, genetic, physiological and anatomical aspects of olfaction are well known in the adult stage, while larval stages olfactory behavior has received some attention it has been less studied than its adult counterpart. Most of these studies focus on olfactory receptor (Or) genes that produce peripheral odor recognition. In this paper, through a loss-of-function screen using P-element inserted lines and also by means of expression analyses of larval olfaction candidate genes, we extended the uncovering of the genetic underpinnings of D. melanogaster larval olfactory behavior by demonstrating that larval olfactory behavior is, in addition to Or genes, orchestrated by numerous genes with diverse functions. Also, our results point out that the genetic architecture of olfactory behavior in D. melanogaster presents a dynamic and changing organization across environments and ontogeny.
Figures


Similar articles
-
Phenotypic plasticity and genotype by environment interaction for olfactory behavior in Drosophila melanogaster.Genetics. 2008 Jun;179(2):1079-88. doi: 10.1534/genetics.108.086769. Epub 2008 May 27. Genetics. 2008. PMID: 18505870 Free PMC article.
-
Dynamic genetic interactions determine odor-guided behavior in Drosophila melanogaster.Genetics. 2006 Nov;174(3):1349-63. doi: 10.1534/genetics.106.060574. Epub 2006 Oct 8. Genetics. 2006. PMID: 17028343 Free PMC article.
-
Functional genomics of odor-guided behavior in Drosophila melanogaster.Chem Senses. 2001 Feb;26(2):215-21. doi: 10.1093/chemse/26.2.215. Chem Senses. 2001. PMID: 11238254
-
Genetic and molecular studies of olfaction in Drosophila.Ciba Found Symp. 1996;200:285-96; discussion 296-301. doi: 10.1002/9780470514948.ch20. Ciba Found Symp. 1996. PMID: 8894304 Review.
-
Molecular genetics of Drosophila olfaction.Ciba Found Symp. 1993;179:150-61; discussion 162-6. Ciba Found Symp. 1993. PMID: 8168375 Review.
Cited by
-
SINE Retrotransposon variation drives Ecotypic disparity in natural populations of Coilia nasus.Mob DNA. 2020 Jan 8;11:4. doi: 10.1186/s13100-019-0198-8. eCollection 2020. Mob DNA. 2020. PMID: 31921363 Free PMC article.
-
The Identification of Congeners and Aliens by Drosophila Larvae.PLoS One. 2015 Aug 27;10(8):e0136363. doi: 10.1371/journal.pone.0136363. eCollection 2015. PLoS One. 2015. PMID: 26313007 Free PMC article.
References
-
- Aceves-Piña EO, Quinn WG. Learning in normal and mutant Drosophila larvae. Science. 1979;206(4414):93–96. - PubMed
-
- Anholt RR, Dilda CL, Chang S, Fanara JJ, Kulkarni NH, Ganguly I, Rollmann SM, Kamdar KP, Mackay TF. The genetic architecture of odor-guided behavior in Drosophila: epistasis and the transcriptome. Nat Genet. 2003;35(2):180–184. - PubMed
-
- Anholt RR, Mackay TF. Quantitative genetic analyses of complex behaviours in Drosophila. Nat Rev Genet. 2004;5(11):838–849. - PubMed
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

