Reactive oxygen species homeostasis and virulence of the fungal pathogen Cryptococcus neoformans requires an intact proline catabolism pathway
- PMID: 23564202
- PMCID: PMC3664852
- DOI: 10.1534/genetics.113.150326
Reactive oxygen species homeostasis and virulence of the fungal pathogen Cryptococcus neoformans requires an intact proline catabolism pathway
Abstract
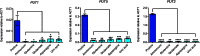
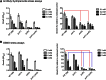
Degradation of the multifunctional amino acid proline is associated with mitochondrial oxidative respiration. The two-step oxidation of proline is catalyzed by proline oxidase and Δ(1)-pyrroline-5-carboxylate (P5C) dehydrogenase, which produce P5C and glutamate, respectively. In animal and plant cells, impairment of P5C dehydrogenase activity results in P5C-proline cycling when exogenous proline is supplied via the actions of proline oxidase and P5C reductase (the enzyme that converts P5C to proline). This proline is oxidized by the proline oxidase-FAD complex that delivers electrons to the electron transport chain and to O2, leading to mitochondrial reactive oxygen species (ROS) overproduction. Coupled activity of proline oxidase and P5C dehydrogenase is therefore important for maintaining ROS homeostasis. In the genome of the fungal pathogen Cryptococcus neoformans, there are two paralogs (PUT1 and PUT5) that encode proline oxidases and a single ortholog (PUT2) that encodes P5C dehydrogenase. Transcription of all three catabolic genes is inducible by the presence of proline. However, through the creation of deletion mutants, only Put5 and Put2 were found to be required for proline utilization. The put2Δ mutant also generates excessive mitochondrial superoxide when exposed to proline. Intracellular accumulation of ROS is a critical feature of cell death; consistent with this fact, the put2Δ mutant exhibits a slight, general growth defect. Furthermore, Put2 is required for optimal production of the major cryptococcal virulence factors. During murine infection, the put2Δ mutant was discovered to be avirulent; this is the first report highlighting the importance of P5C dehydrogenase in enabling pathogenesis of a microorganism.
Keywords: Cryptococcus neoformans; P5C dehydrogenase; nitrogen; proline oxidase; virulence.
Figures





References
-
- Altschul S. F., Gish W., Miller W., Myers E. W., Lipman D. J., 1990. Basic local alignment search tool. J. Mol. Biol. 215: 403–410. - PubMed
-
- Arst H. N., Jr, MacDonald D. W., 1978. Reduced expression of a distal gene of the prn gene cluster in deletion mutants of Aspergillus nidulans: genetic evidence for a dicistronic messenger in an eukaryote. Mol. Gen. Genet. 163: 17–22. - PubMed
-
- Atkinson D. E., 1977. Cellular Energy Metabolism and Its Regulation. Academic Press, New York.
-
- Brachmann C. B., Davies A., Cost G. J., Caputo E., Li J., et al. , 1998. Designer deletion strains derived from Saccharomyces cerevisiae S288C: a useful set of strains and plasmids for PCR-mediated gene disruption and other applications. Yeast 14: 115–132. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

