Genome of the pathogen Porphyromonas gingivalis recovered from a biofilm in a hospital sink using a high-throughput single-cell genomics platform
- PMID: 23564253
- PMCID: PMC3638142
- DOI: 10.1101/gr.150433.112
Genome of the pathogen Porphyromonas gingivalis recovered from a biofilm in a hospital sink using a high-throughput single-cell genomics platform
Abstract
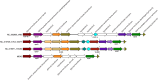
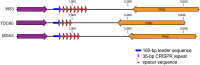
Although biofilms have been shown to be reservoirs of pathogens, our knowledge of the microbial diversity in biofilms within critical areas, such as health care facilities, is limited. Available methods for pathogen identification and strain typing have some inherent restrictions. In particular, culturing will yield only a fraction of the species present, PCR of virulence or marker genes is mainly focused on a handful of known species, and shotgun metagenomics is limited in the ability to detect strain variations. In this study, we present a single-cell genome sequencing approach to address these limitations and demonstrate it by specifically targeting bacterial cells within a complex biofilm from a hospital bathroom sink drain. A newly developed, automated platform was used to generate genomic DNA by the multiple displacement amplification (MDA) technique from hundreds of single cells in parallel. MDA reactions were screened and classified by 16S rRNA gene PCR sequence, which revealed a broad range of bacteria covering 25 different genera representing environmental species, human commensals, and opportunistic human pathogens. Here we focus on the recovery of a nearly complete genome representing a novel strain of the periodontal pathogen Porphyromonas gingivalis (P. gingivalis JCVI SC001) using the single-cell assembly tool SPAdes. Single-cell genomics is becoming an accepted method to capture novel genomes, primarily in the marine and soil environments. Here we show for the first time that it also enables comparative genomic analysis of strain variation in a pathogen captured from complex biofilm samples in a healthcare facility.
Figures



References
-
- Badger JH, Hoover TR, Brun YV, Weiner RM, Laub MT, Alexandre G, Mrazek J, Ren Q, Paulsen IT, Nelson KE, et al. 2006. Comparative genomic evidence for a close relationship between the dimorphic prosthecate bacteria Hyphomonas neptunium and Caulobacter crescentus. J Bacteriol 188: 6841–6850 - PMC - PubMed
-
- Barrangou R, Fremaux C, Deveau H, Richards M, Boyaval P, Moineau S, Romero DA, Horvath P 2007. CRISPR provides acquired resistance against viruses in prokaryotes. Science 315: 1709–1712 - PubMed
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
