Transcriptional corepressors HIPK1 and HIPK2 control angiogenesis via TGF-β-TAK1-dependent mechanism
- PMID: 23565059
- PMCID: PMC3614511
- DOI: 10.1371/journal.pbio.1001527
Transcriptional corepressors HIPK1 and HIPK2 control angiogenesis via TGF-β-TAK1-dependent mechanism
Abstract
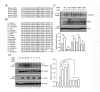
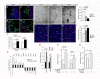
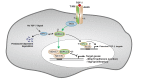
Several critical events dictate the successful establishment of nascent vasculature in yolk sac and in the developing embryos. These include aggregation of angioblasts to form the primitive vascular plexus, followed by the proliferation, differentiation, migration, and coalescence of endothelial cells. Although transforming growth factor-β (TGF-β) is known to regulate various aspects of vascular development, the signaling mechanism of TGF-β remains unclear. Here we show that homeodomain interacting protein kinases, HIPK1 and HIPK2, are transcriptional corepressors that regulate TGF-β-dependent angiogenesis during embryonic development. Loss of HIPK1 and HIPK2 leads to marked up-regulations of several potent angiogenic genes, including Mmp10 and Vegf, which result in excessive endothelial proliferation and poor adherens junction formation. This robust phenotype can be recapitulated by siRNA knockdown of Hipk1 and Hipk2 in human umbilical vein endothelial cells, as well as in endothelial cell-specific TGF-β type II receptor (TβRII) conditional mutants. The effects of HIPK proteins are mediated through its interaction with MEF2C, and this interaction can be further enhanced by TGF-β in a TAK1-dependent manner. Remarkably, TGF-β-TAK1 signaling activates HIPK2 by phosphorylating a highly conserved tyrosine residue Y-361 within the kinase domain. Point mutation in this tyrosine completely eliminates the effect of HIPK2 as a transcriptional corepressor in luciferase assays. Our results reveal a previously unrecognized role of HIPK proteins in connecting TGF-β signaling pathway with the transcriptional programs critical for angiogenesis in early embryonic development.
Conflict of interest statement
The authors have declared that no competing interests exist.
Figures





References
-
- Carmeliet P (2005) Angiogenesis in life, disease and medicine. Nature 438: 932–936. - PubMed
-
- Rossant J, Howard L (2002) Signaling pathways in vascular development. Annual Review of Cell and Developmental Biology 18: 541–573. - PubMed
-
- Coultas L, Chawengsaksophak K, Rossant J (2005) Endothelial cells and VEGF in vascular development. Nature 438: 937–945. - PubMed
-
- You LR, Lin FJ, Lee CT, DeMayo FJ, Tsai MJ, et al. (2005) Suppression of Notch signalling by the COUP-TFII transcription factor regulates vein identity. Nature 435: 98–104. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Miscellaneous

