Simultaneous and independent detection of C9ORF72 alleles with low and high number of GGGGCC repeats using an optimised protocol of Southern blot hybridisation
- PMID: 23566336
- PMCID: PMC3626718
- DOI: 10.1186/1750-1326-8-12
Simultaneous and independent detection of C9ORF72 alleles with low and high number of GGGGCC repeats using an optimised protocol of Southern blot hybridisation
Abstract
Background: Sizing of GGGGCC hexanucleotide repeat expansions within the C9ORF72 locus, which account for approximately 10% of all amyotrophic lateral sclerosis (ALS) cases, is urgently required to answer fundamental questions about mechanisms of pathogenesis in this important genetic variant. Currently employed PCR protocols are limited to discrimination between the presence and absence of a modified allele with more than 30 copies of the repeat, while Southern hybridisation-based methods are confounded by the somatic heterogeneity commonly present in blood samples, which might cause false-negative or ambiguous results.
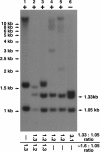
Results: We describe an optimised Southern hybridisation-based protocol that allows confident detection of the presence of a C9ORF72 repeat expansion alongside independent assessment of its heterogeneity and the number of repeat units. The protocol can be used with either a radiolabeled or non-radiolabeled probe. Using this method we have successfully sized the C9ORF72 repeat expansion in lymphoblastoid cells, peripheral blood, and post-mortem central nervous system (CNS) tissue from ALS patients. It was also possible to confidently demonstrate the presence of repeat expansion, although of different magnitude, in both C9ORF72 alleles of the genome of one patient.
Conclusions: The suggested protocol has sufficient advantages to warrant adoption as a standard for Southern blot hybridisation analysis of GGGGCC repeat expansions in the C9ORF72 locus.
Figures


References
-
- DeJesus-Hernandez M, Mackenzie, Ian R, Boeve, Bradley F, Boxer, Adam L, Baker M, Rutherford, Nicola J, Nicholson, Alexandra M, Finch, NiCole A, Flynn H, Adamson J, Kouri N, Wojtas A, Sengdy P, Hsiung G, Yuek R, Karydas A, Seeley, William W, Josephs, Keith A, Coppola G, Geschwind, Daniel H, Wszolek, Zbigniew K, Feldman H, Knopman, David S, Petersen, Ronald C, Miller, Bruce L, Dickson, Dennis W, Boylan, Kevin B, Graff-Radford, Neill R, Rademakers R. Expanded GGGGCC hexanucleotide repeat in noncoding region of C9ORF72 causes chromosome 9p-linked FTD and ALS. Neuron. 2011;72:245–256. doi: 10.1016/j.neuron.2011.09.011. - DOI - PMC - PubMed
-
- Renton, Alan E, Majounie E, Waite A, Simón-Sánchez J, Rollinson S, Gibbs JR, Schymick, Jennifer C, Laaksovirta H, Van Swieten, John C, Myllykangas L, Kalimo H, Paetau A, Abramzon Y, Remes, Anne M, Kaganovich A, Scholz, Sonja W, Duckworth J, Ding J, Harmer, Daniel W, Hernandez, Dena G, Johnson, Janel O, Mok K, Ryten M, Trabzuni D, Guerreiro, Rita J, Orrell, Richard W, Neal J, Murray A, Pearson J, Jansen, Iris E. A Hexanucleotide Repeat Expansion in C9ORF72 Is the Cause of Chromosome 9p21-Linked ALS-FTD. Neuron. 2011;72:257–268. doi: 10.1016/j.neuron.2011.09.010. - DOI - PMC - PubMed
-
- Ash PE, Bieniek KF, Gendron TF, Caulfield T, Lin WL, DeJesus-Hernandez M, van Blitterswijk MM, Jansen-West K, Paul JW 3rd, Rademakers R, Boylan KB, Dickson DW, Petrucelli L. Unconventional Translation of C9ORF72 GGGGCC Expansion Generates Insoluble Polypeptides Specific to c9FTD/ALS. Neuron. 2013;77:639–646. doi: 10.1016/j.neuron.2013.02.004. - DOI - PMC - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Miscellaneous

