Liposome delivery of microRNA-145 to mesenchymal stem cells leads to immunological off-target effects mediated by RIG-I
- PMID: 23568258
- PMCID: PMC3677300
- DOI: 10.1038/mt.2013.55
Liposome delivery of microRNA-145 to mesenchymal stem cells leads to immunological off-target effects mediated by RIG-I
Abstract
Synthetic microRNAs regulate gene expression when transfected into cells, and may be used in strategies for molecular therapy both in vitro and in vivo. Liposomal transfection reagents are frequently used as delivery vehicles in both settings. Here, we report on the immunological off-target effects observed following liposome transfection of synthetic microRNA-145 into human mesenchymal stem cells and human articular chondrocytes (hAC). The immune response was independent on endosome delivery and toll-like receptors (TLRs) but was mediated by retinoic acid inducible-gene 1 (RIG-I). Upregulation of immune genes required liposomal delivery, as no immune response was observed after electroporation of smiR-145 directly in to the cytosol, suggesting a new role of RIG-I. Immune response was seen both with blunt ended and 2-nucleotide 3' overhang versions of synthetic miR-145, and occurred in the absence of a 5'ppp cap. Mutations in a centrally placed poly (UUUU) sequence reduced, but did not abolish the immune response. Interestingly, exposure to liposomes alone led to upregulation of several immune genes, including RIG-I mRNA. However, this process was not mediated by RIG-I. This insight is important for researchers to avoid unexpected results from gene transfer experiments in vitro and unwanted immune responses following the use of lipid-based transfection reagents in vivo.
Figures
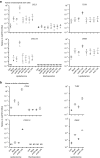




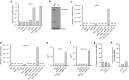

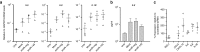
References
-
- Hutvágner G, Zamore PD. A microRNA in a multiple-turnover RNAi enzyme complex. Science. 2002;297:2056–2060. - PubMed
-
- Lee RC, Feinbaum RL, Ambros V. The C. elegans heterochronic gene lin-4 encodes small RNAs with antisense complementarity to lin-14. Cell. 1993;75:843–854. - PubMed
-
- Vasudevan S. Posttranscriptional upregulation by microRNAs. Wiley Interdiscip Rev RNA. 2012;3:311–330. - PubMed
-
- Hornung V, Guenthner-Biller M, Bourquin C, Ablasser A, Schlee M, Uematsu S, et al. Sequence-specific potent induction of IFN-alpha by short interfering RNA in plasmacytoid dendritic cells through TLR7. Nat Med. 2005;11:263–270. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Miscellaneous

